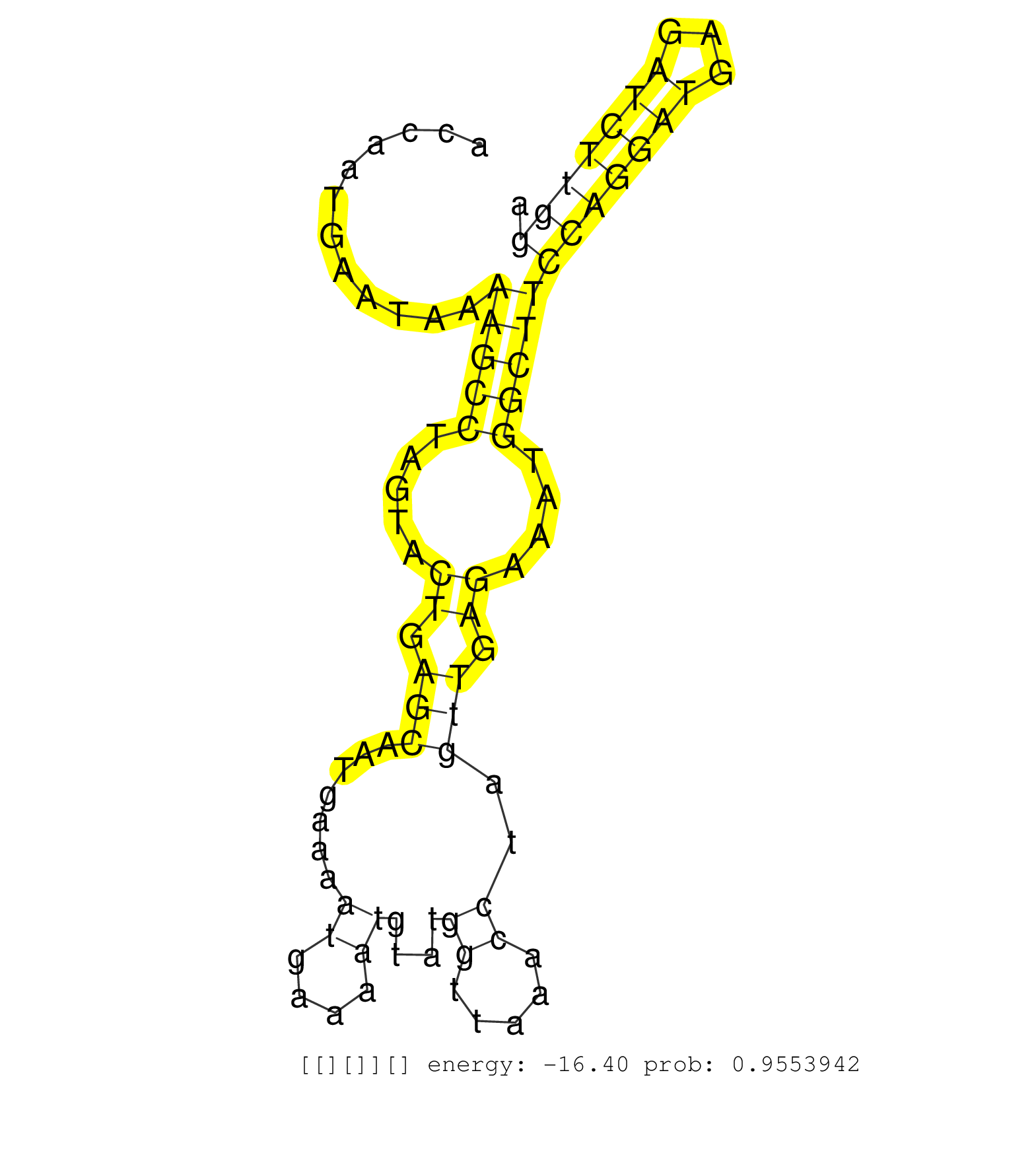

| Gene: Zfyve19 | ID: uc008lti.1_intron_5_0_chr2_119037751_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| GCAAAGTGAGTTACAGGCAGGCCAGGACTACACAGAGAAACCTTGCCTTGAAAAAAACAAAAATAAATACACCAATGAATAAAAGCCTAGTACTGAGCAATGAAAATGAAAATGTATGGTTAAACCTAGTTGAGAAATGGCTTCCAGGATGAGATCTTGGACTTCTTCTGTCTTCCTAACTACATCACCCCTGCCAGCAGCCTCTCTCCAGAATGACCTCAACAAGGGAGCTGCGAGAAGCCAGCGCACA ..................................................................................(((((.....((.(((.......((....))....((.....))..))).))....)))))(((((((...))))))).......................................................................................... ......................................................................71........................................................................................161....................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................TGAATAAAAGCCTAGTACTGAGCAAT..................................................................................................................................................... | 26 | 1 | 9.00 | 9.00 | 3.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................TGAATAAAAGCCTAGTACTGAGCAATG.................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................TGAATAAAAGCCTAGTACTGAGCAATGA................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGACCTCAACAAGGGAGCTGCGAGAA........... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGAGAAATGGCTTCCAGGATGAGATCT............................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGGTTAAACCTAGTTGAGAAATGGCTTC.......................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TGAGTTACAGGCAGGCCAGGACTACA.......................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TCTCCAGAATGACCTCAACAAGGGAGCT.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................AGAATGACCTCAACAAGGGAGCTGCGA.............. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCCAGAATGACCTCAACAAGGGAGCT.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TGGTTAAACCTAGTTGAGAAATGGCTT........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................GAATGACCTCAACAAGGGAGCTGCGAGA............ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TCCTAACTACATCACtgc........................................................... | 18 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................TGGTTAAACCTAGTTGAGAAATGGCTTt.......................................................................................................... | 28 | t | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGAGATCTTGGACTTCTTCTGTCTTC........................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................TAGTTGAGAAATGGCTTCCAGGATGA.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCCAGAATGACCTCAACAAGGGAGCTGC................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACCTAGTTGAGAAATGGCTTCCAGG...................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TGAATAAAAGCCTAGTACTGAGCAA...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGAGATCTTGGACTTCTTCTGTCTTCCTAAC...................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGCCAGCAGCCTCTCTCCAGAATGACC................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................TGAATAAAAGCCTAGTACTGAGCAATGAA.................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGACCTCAACAAGGGAGCTGCGAGAAa.......... | 27 | a | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TACACAGAGAAACCTTGCCTTGAAAAAA.................................................................................................................................................................................................. | 28 | 5 | 0.60 | 0.60 | - | - | - | - | - | - | - | 0.20 | - | 0.20 | - | - | - | - | - | - | 0.20 | - |
| ..............................CACAGAGAAACCTTGCCTTGAAAAAA.................................................................................................................................................................................................. | 26 | 6 | 0.33 | 0.33 | - | 0.17 | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - |
| .................CAGGCCAGGACTACACAGAG..................................................................................................................................................................................................................... | 20 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - |
| ............................TACACAGAGAAACCTTGCCTTGAAAAA................................................................................................................................................................................................... | 27 | 12 | 0.17 | 0.17 | - | - | - | - | - | - | - | 0.08 | - | - | 0.08 | - | - | - | - | - | - | - |
| ..............................CACAGAGAAACCTTGCCTTGAAAAA................................................................................................................................................................................................... | 25 | 13 | 0.08 | 0.08 | - | - | - | - | - | - | - | 0.08 | - | - | - | - | - | - | - | - | - | - |
| ...........................CTACACAGAGAAACCTTGCCTTGAAAA.................................................................................................................................................................................................... | 27 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - | - | - | - |
| ............................TACACAGAGAAACCTTGCCTTGAAAA.................................................................................................................................................................................................... | 26 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - |
| ...............................ACAGAGAAACCTTGCCTTGAAAAA................................................................................................................................................................................................... | 24 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 |
| GCAAAGTGAGTTACAGGCAGGCCAGGACTACACAGAGAAACCTTGCCTTGAAAAAAACAAAAATAAATACACCAATGAATAAAAGCCTAGTACTGAGCAATGAAAATGAAAATGTATGGTTAAACCTAGTTGAGAAATGGCTTCCAGGATGAGATCTTGGACTTCTTCTGTCTTCCTAACTACATCACCCCTGCCAGCAGCCTCTCTCCAGAATGACCTCAACAAGGGAGCTGCGAGAAGCCAGCGCACA ..................................................................................(((((.....((.(((.......((....))....((.....))..))).))....)))))(((((((...))))))).......................................................................................... ......................................................................71........................................................................................161....................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................GCCAGGACTACACAGAGAaaa.................................................................................................................................................................................................................... | 21 | aaa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |