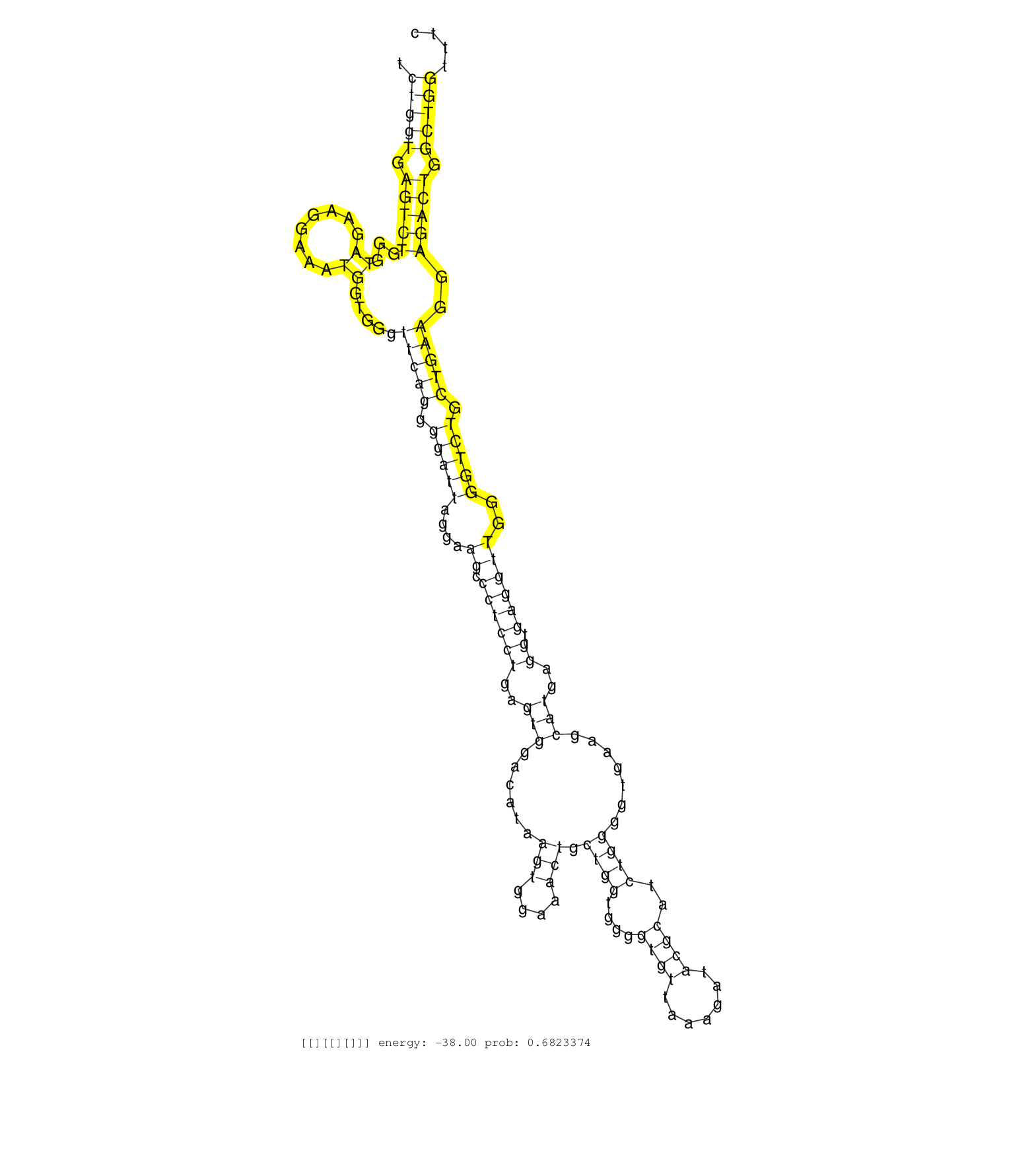
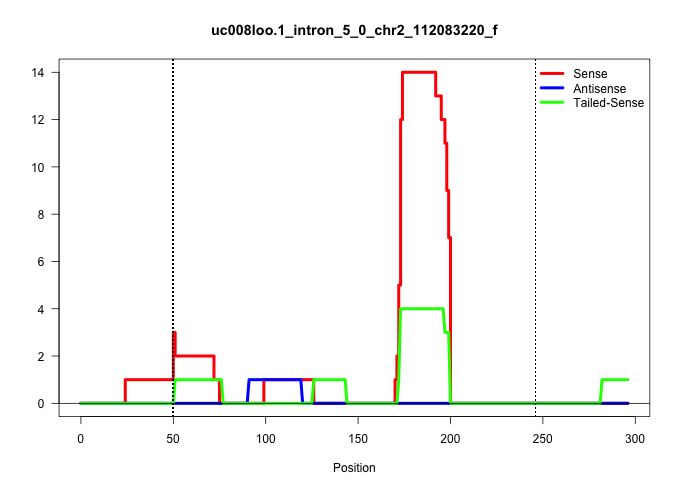
| Gene: Agpat7 | ID: uc008loo.1_intron_5_0_chr2_112083220_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| TCGCAGGGGTGCCCGTGCAGCCCGTCCTCATCCGATACCCCAACAGTCTGGTGAGTCTGGGTAGAAGGAAATGGTGGGTTCAGGGGATTAGGAAGCCCTCCTGAGTGGACATAAGTGGAAACTGCTGGTGGGGTGTTAAAGATACGCATCTGGGGTGAAGCATGAGGTGAGGTTGGGGTCTGCTGAAGGAGACTGGCTGGTTTCCAGGAAGAGAACTAACCTCTGCCCCTTCTGCTTTTCCCACAGGACACCACCAGCTGGGCATGGAGGGGCCCCGGCGTGTGAGTGCTAGGGTC ...............................................(((((.(((((...((........)).....(((((.(((((....((.((((((..(((......(((....))).((((....((((.......))))..)))).......)))..)).))))))..))))).)))))..))))).)))))................................................................................................ ..............................................47...........................................................................................................................................................204.......................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACTGGCTGG................................................................................................ | 27 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGTCTGGGTAGAAGGAAATGGTa............................................................................................................................................................................................................................ | 26 | a | 3.00 | 1.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TTGGGGTCTGCTGAAGGAGACTGGCT.................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................GGGGTCTGCTGAAGGAGACTGGCTGG................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACTGGCTG................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................GTTGGGGTCTGCTGAAGGAGACTGGC................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTCTGGGTAGAAGGAAATGGTGt........................................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GGTTGGGGTCTGCTGAAGGAGA........................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGAACTAACCTCTGCCCCTTCTGCTTTTCCCACAG.................................................. | 35 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................TCCTCATCCGATACCCCAACAGTCTGG..................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGAGTCTGGGTAGAAGGAAAT................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACTGGCTtt................................................................................................ | 27 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CCTGAGTGGACATAAGTGGAAACTGCT.......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TTGGGGTCTGCTGAAGGAGACTGGt................................................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACTG..................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACTGGtaat................................................................................................ | 27 | taat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACT...................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................TGGGGTCTGCTGAAGGAGACTGGCTGa................................................................................................ | 27 | a | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................GGTGGGGTGTTAAAaaaa........................................................................................................................................................ | 18 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TTGGGGTCTGCTGAAGGAGACTGGCTGG................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGAGTCTGGGTAGAAGGAAATGGT............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................TGAGTGCTAGGGTCgggg | 18 | gggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| TCGCAGGGGTGCCCGTGCAGCCCGTCCTCATCCGATACCCCAACAGTCTGGTGAGTCTGGGTAGAAGGAAATGGTGGGTTCAGGGGATTAGGAAGCCCTCCTGAGTGGACATAAGTGGAAACTGCTGGTGGGGTGTTAAAGATACGCATCTGGGGTGAAGCATGAGGTGAGGTTGGGGTCTGCTGAAGGAGACTGGCTGGTTTCCAGGAAGAGAACTAACCTCTGCCCCTTCTGCTTTTCCCACAGGACACCACCAGCTGGGCATGGAGGGGCCCCGGCGTGTGAGTGCTAGGGTC ...............................................(((((.(((((...((........)).....(((((.(((((....((.((((((..(((......(((....))).((((....((((.......))))..)))).......)))..)).))))))..))))).)))))..))))).)))))................................................................................................ ..............................................47...........................................................................................................................................................204.......................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................GAAGCCCTCCTGAGTGGACATAAGTGGAA................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |