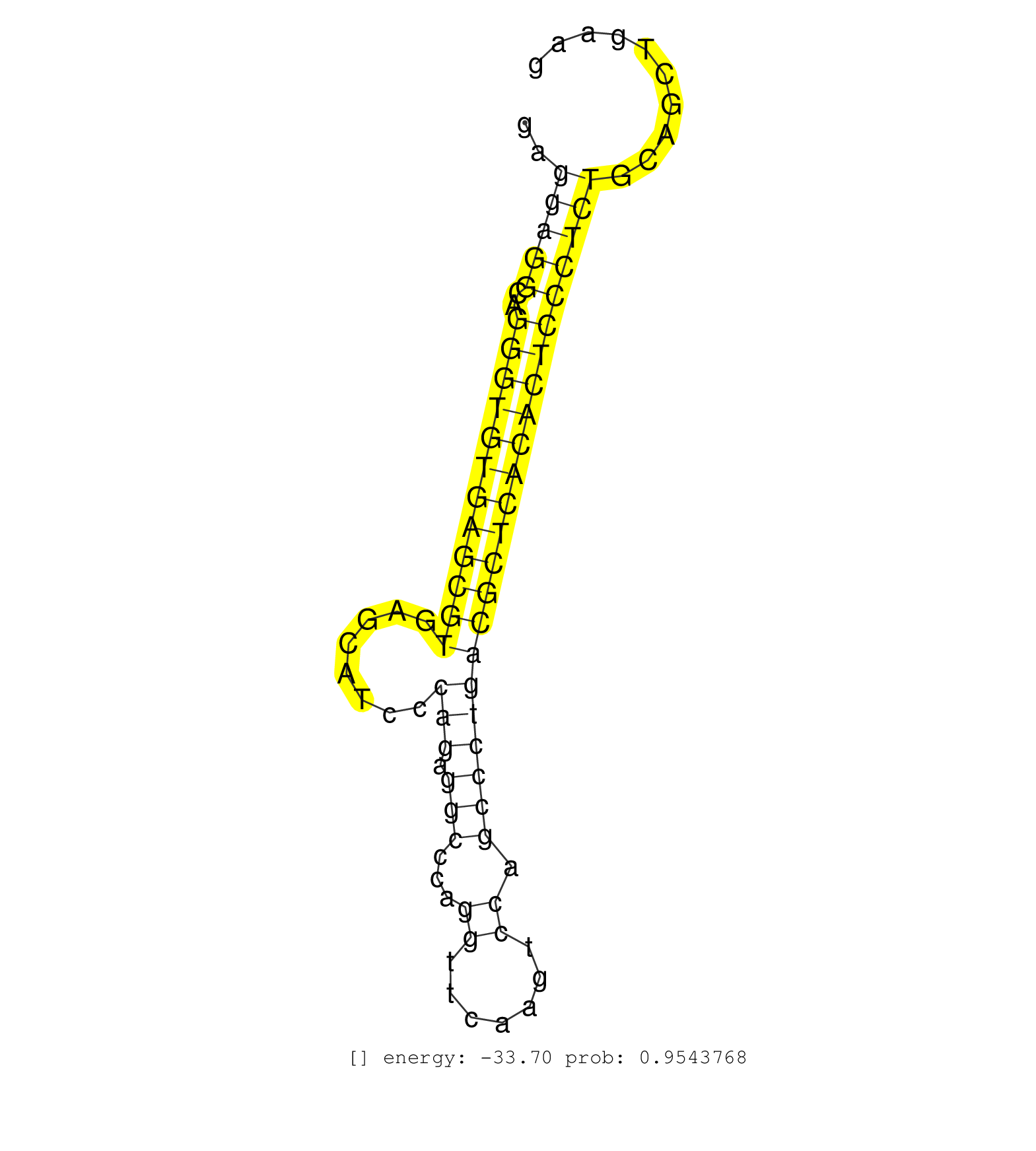

| Gene: Cd82 | ID: uc008lfx.1_intron_3_0_chr2_93262086_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| AAAAATGCCTGCCGAAGGGTAGGCCATGGCTGACAGGGTTGGCGCTGAGCAAATGGGCACTAGGGATCACCAGGACAGTGGATGTTCTGCAGCCATGAGCGTGTCCTGGGATTGCTGTGGTGGAGGAGGCAGGGTGTGAGCGTGAGCATCCCAGAGGCCCAGGTTCAAGTCCAGCCCTGACGCTCACACTCCCTCTGCAGCTGAAGAAGGAGATGGGGAACACAGTGATGGACATCATTCGCAACTACAC ............................................................................................................................(((((..((((((((((((........(((.(((...((.......)).)))))))))))))))))))))))...................................................... ..........................................................................................................................123................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................CGCTCACACTCCCTCTGCAGtt................................................ | 22 | tt | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GAAGAAGGAGATGGGGAACACAGT........................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GGCAGGGTGTGAGCGTGAGCAT..................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................TGAAGAAGGAGATGGGGAACACAGTGATGt................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................................................................................................TGGGGAACACAGTGATGGACATCATTC.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGCTGAAGAAGGAGATGGGGAAC............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TGGACATCATTCGCAACT.... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CGCTCACACTCCCTCTGCAGa................................................. | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGGGATTGCTGTGGTGGAGGAGGCAGt..................................................................................................................... | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CGTGTCCTGGGATTcgg...................................................................................................................................... | 17 | cgg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TGTTCTGCAGCCATGAGCGTGTCCTGt............................................................................................................................................. | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGCGTGTCCTGGGAaatt....................................................................................................................................... | 18 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................CGCTCACACTCCCTCTGCAGaa................................................ | 22 | aa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGAGCGTGTCCTGGGATTGCTGTGGT................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................TGGGATTGCTGTGGTGGAGGAGGg........................................................................................................................ | 24 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AAAAATGCCTGCCGAAGGGTAGGCCATGGCTGACAGGGTTGGCGCTGAGCAAATGGGCACTAGGGATCACCAGGACAGTGGATGTTCTGCAGCCATGAGCGTGTCCTGGGATTGCTGTGGTGGAGGAGGCAGGGTGTGAGCGTGAGCATCCCAGAGGCCCAGGTTCAAGTCCAGCCCTGACGCTCACACTCCCTCTGCAGCTGAAGAAGGAGATGGGGAACACAGTGATGGACATCATTCGCAACTACAC ............................................................................................................................(((((..((((((((((((........(((.(((...((.......)).)))))))))))))))))))))))...................................................... ..........................................................................................................................123................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................CCCAGGTTCAAGTaggc................................................................................ | 17 | aggc | 5.00 | 0.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................CCCAGGTTCAAGTaagc................................................................................ | 17 | aagc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGGTGGAGGAGGCAGtacc...................................................................................................................... | 19 | tacc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |