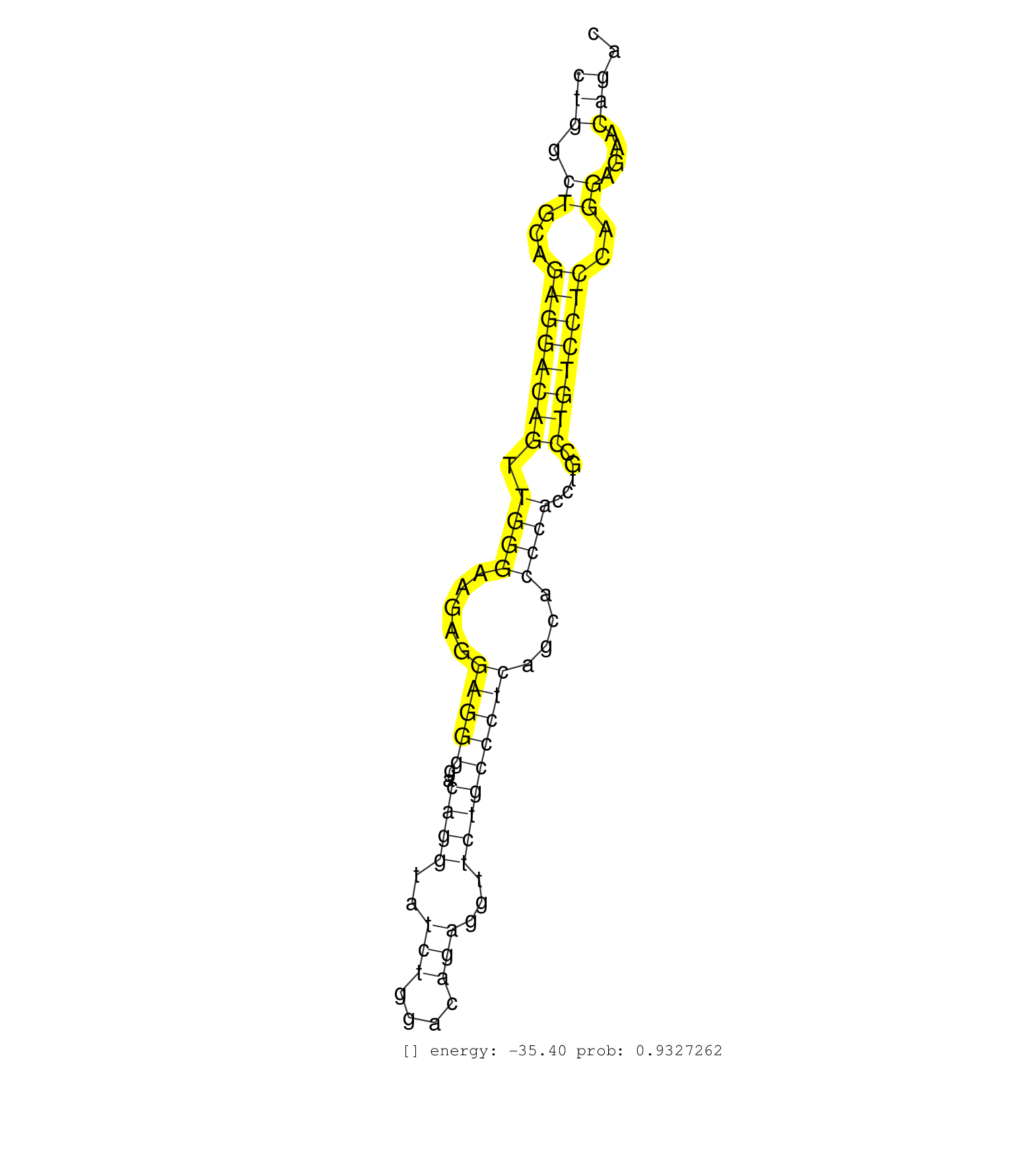

| Gene: Mapk8ip1 | ID: uc008kxv.1_intron_7_0_chr2_92227541_r | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(22) TESTES |
| GCACAGTTGGCAGGACCGTGTGTCTCGATCATCCTCCCCTCTGAAGACAGGTAAGTCAGGCCTCTCTCCCCAAGCTGGAGACCCAGCAGTCTTCACCTGGCTCACCTGGGCTGAGGTCATCTGGAAGCCAGTGGCTGGCTGCAGAGGACAGTTGGGAAGAGGAGGGGACAGGTATCTGGACAGAGGTTCTGCCCTCAGCACCCACCTGCCTGTCCTCCAGGAGAACAGACGCCTCCACATGAACACATCTGCCTGAGTGATGAGCTGCCA ......................................................................................................................................(((.((...((((((((.((((.....(((((..((((..(((....)))...)))))))))....)))).....))))))))..))....))).......................................... ......................................................................................................................................135............................................................................................230...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................................TGAACACATCTGCCTGAGTGATGAGCTG... | 28 | 1 | 9.00 | 9.00 | - | 2.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGCCTGTCCTCCAGGAGctca........................................... | 21 | ctca | 7.00 | 0.00 | 6.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AGGGGACAGGTATCTGGACAGA...................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................TGCAGAGGACAGTTGGGAAGAGcagt......................................................................................................... | 26 | cagt | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GCCTGTCCTCCAGGAGctc............................................ | 19 | ctc | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ACAGTTGGCAGGACatta.......................................................................................................................................................................................................................................................... | 18 | atta | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CCTCTGAAGACAGGTAAGTCA.................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................TGAACACATCTGCCTGAGTGATGAGCTGC.. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................ATGAACACATCTGCCTGAGTGATGAGC..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................TGCAGAGGACAGTTGGGAAGAGcaga......................................................................................................... | 26 | caga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AGGAGGGGACAGGTATCTGGACAG....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................................................TGAACACATCTGCCTGAGTGATGAGC..... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGCTGCAGAGGACAGTTGGGAAGA.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................CACATGAACACATCTGCCTGAGTGATG........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................TGAACACATCTGCCTGAGTGATGAGCT.... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AGGGGACAGGTATCTGGACAGAGGTTCT................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................TCTCGATCATCCTCCCCTCTGAAGACAGG........................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGCCTGTCCTCCAGGAGctc............................................ | 20 | ctc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGCAGAGGACAGTTGGGAAGAGcata......................................................................................................... | 26 | cata | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AGGGGACAGGTATCTGGACAGAGG.................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................................................................................AGAACAGACGCCTCCACATGAACA......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................AAGCCAGTGGCTGGCTGCAGAGGACAGT...................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| GCACAGTTGGCAGGACCGTGTGTCTCGATCATCCTCCCCTCTGAAGACAGGTAAGTCAGGCCTCTCTCCCCAAGCTGGAGACCCAGCAGTCTTCACCTGGCTCACCTGGGCTGAGGTCATCTGGAAGCCAGTGGCTGGCTGCAGAGGACAGTTGGGAAGAGGAGGGGACAGGTATCTGGACAGAGGTTCTGCCCTCAGCACCCACCTGCCTGTCCTCCAGGAGAACAGACGCCTCCACATGAACACATCTGCCTGAGTGATGAGCTGCCA ......................................................................................................................................(((.((...((((((((.((((.....(((((..((((..(((....)))...)))))))))....)))).....))))))))..))....))).......................................... ......................................................................................................................................135............................................................................................230...................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|