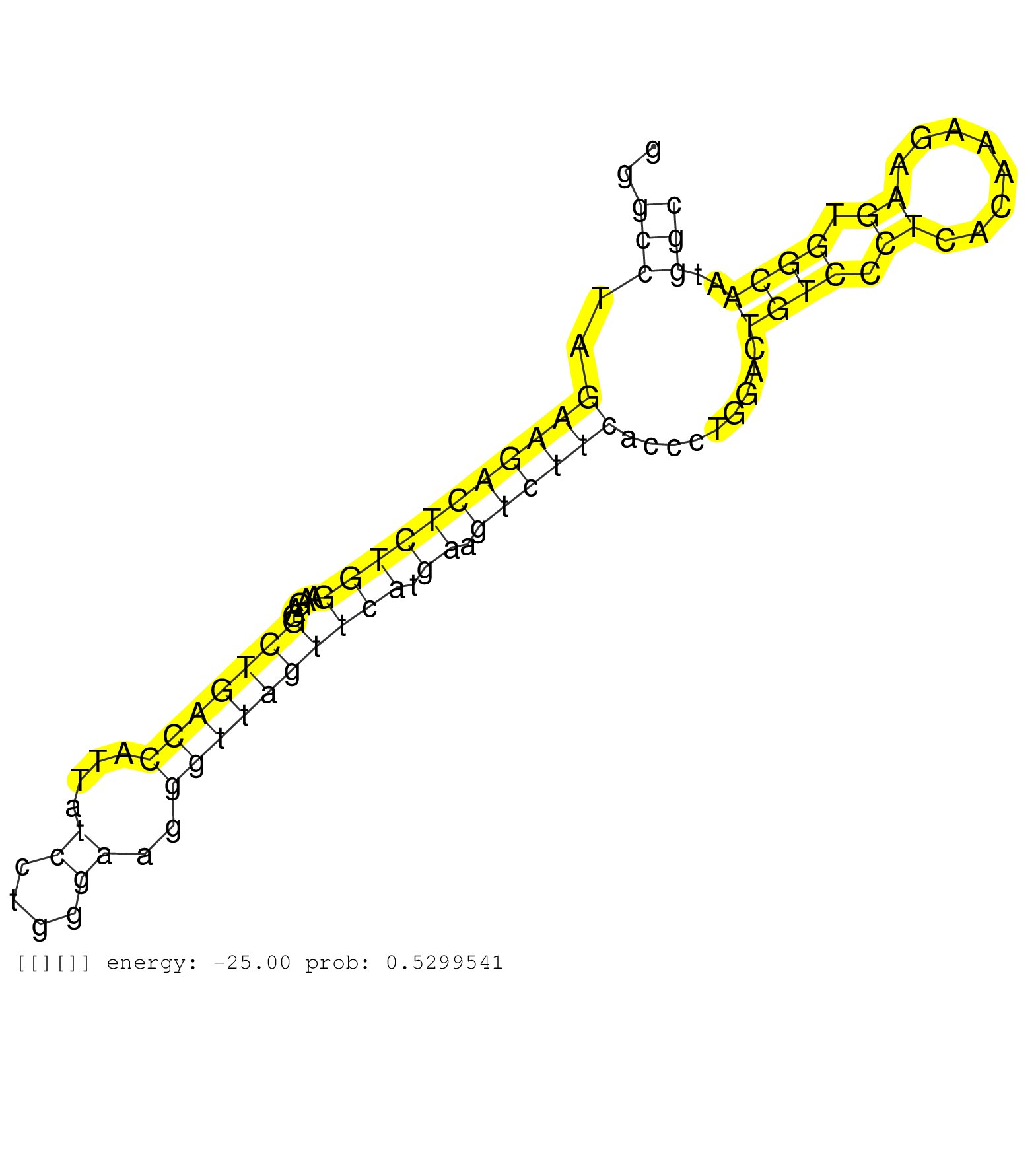

| Gene: 1700029I15Rik | ID: uc008kxu.1_intron_0_0_chr2_92223209_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| CTTTTTTACCTCATCTTGGCAGGCGCACTCCTTCGACCTCAGCCCCAGAGGTAAGGCCAGAGCCTCAGGGCCTAGAAGACTCTGGAAGAGGCTGACCATTATCCTGGGAAGGGTTAGTTCATGAAGTCTTCACCCTGGACTGTCCCTCACAAAGAAGTGGCAATGGCCTTTTGGGTTAGGGCTCAGCACAGCAATATTTCCCATAGGTCTCAGCAATCTGTTCCGGAGGAATTTTCAGCCCCGCTGGAACTGTTGC .....................................................................(((..(((((((((((.....(((((((....((....))..)))))))))).)).)))))).........((((.((........)).))))..)))......................................................................................... ...................................................................68.................................................................................................167....................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TAGAAGACTCTGGAAGAGGCTGACCATT............................................................................................................................................................ | 28 | 1 | 12.00 | 12.00 | 3.00 | 6.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGACTGTCCCTCACAAAGAAGTGGCAA............................................................................................. | 28 | 1 | 7.00 | 7.00 | - | - | 2.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................................TAGGTCTCAGCAATCTGTTCCGGAGG........................... | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................TAGAAGACTCTGGAAGAGGCTGACCAT............................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TAGAAGACTCTGGAAGAGGCTGACCA.............................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTCCCTCACAAAGAAGTGGCAATGGCCTT...................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................TGGACTGTCCCTCACAAAGAAGTGGCAAT............................................................................................ | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GAAGACTCTGGAAGAGGCTGACCATTA........................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TGAAGTCTTCACCCTGGACT................................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CTGGACTGTCCCTCACAAAGAAGTGGCAAT............................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TAGAAGACTCTGGAAGAGGCTGACCATTA........................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................CAGCACAGCAATATTggc....................................................... | 18 | ggc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CACAAAGAAGTGGCAATGGCCTTTTGGGT................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................AGAAGACTCTGGAAGAGGCTGACCATTA........................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGACTGTCCCTCACAAAGAAGTGGCA.............................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TAGAAGACTCTGGAAGAGGCTGACC............................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................AAGACTCTGGAAGAGGCTGACCATT............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CTGGACTGTCCCTCACAAAGAAGTGGC............................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGGGCCTAGAAGACTCTGGAAGAGGCT................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TAGAAGACTCTGGAAGAGGCTGACCATTtt.......................................................................................................................................................... | 30 | tt | 1.00 | 12.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................CCTAGAAGACTCTGGAAGAGGCTGACCAT............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TCTGGAAGAGGCTGACCATTATCCTGGt.................................................................................................................................................... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................CATGAAGTCTTCACCCTGGACTGTCCC.............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................ATTTCCCATAGGTCTCAGCAATCTGTTC................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................AGAAGACTCTGGAAGAGGCTGACCATT............................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................TGGAAGAGGCTGACCATTATCCTGGGA................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AAGACTCTGGAAGAGGCTGACCATTAT.......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CTTTTTTACCTCATCTTGGCAGGCGCACTCCTTCGACCTCAGCCCCAGAGGTAAGGCCAGAGCCTCAGGGCCTAGAAGACTCTGGAAGAGGCTGACCATTATCCTGGGAAGGGTTAGTTCATGAAGTCTTCACCCTGGACTGTCCCTCACAAAGAAGTGGCAATGGCCTTTTGGGTTAGGGCTCAGCACAGCAATATTTCCCATAGGTCTCAGCAATCTGTTCCGGAGGAATTTTCAGCCCCGCTGGAACTGTTGC .....................................................................(((..(((((((((((.....(((((((....((....))..)))))))))).)).)))))).........((((.((........)).))))..)))......................................................................................... ...................................................................68.................................................................................................167....................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................TCAGCAATCTGTTCCGGAGGAATT....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |