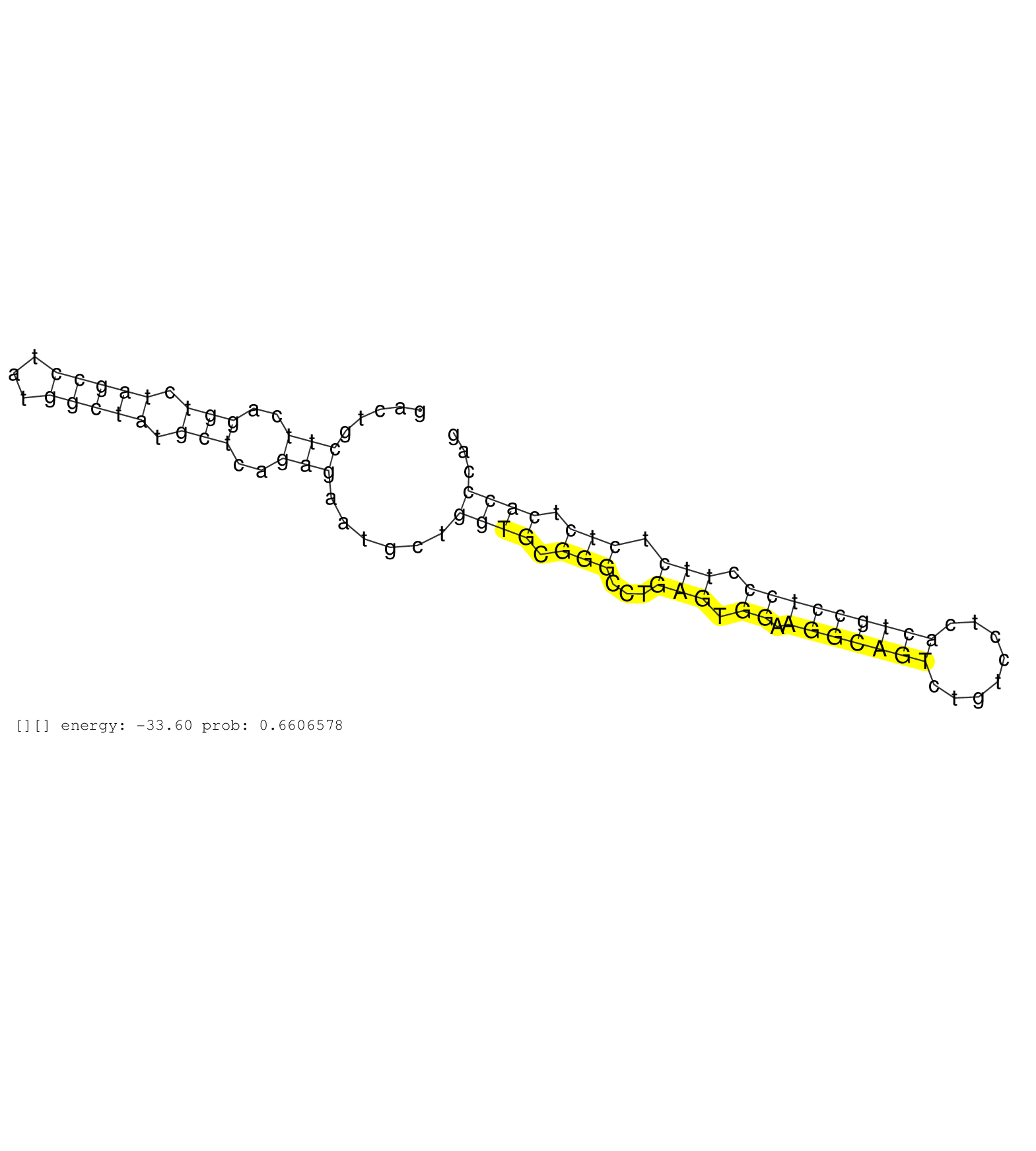
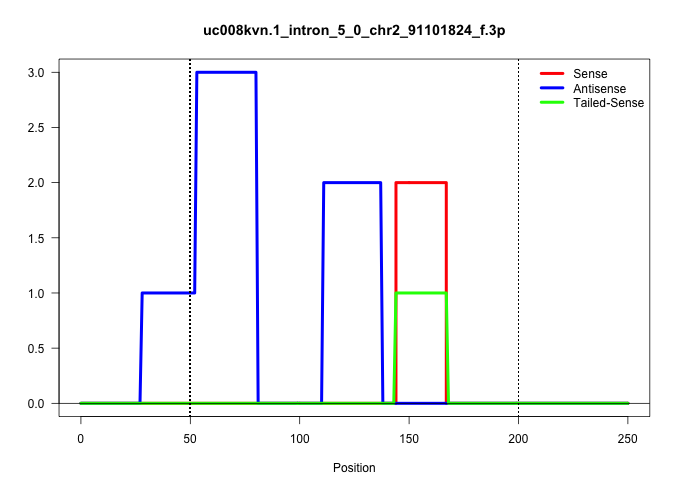
| Gene: Pacsin3 | ID: uc008kvn.1_intron_5_0_chr2_91101824_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(13) TESTES |
| TGCACTTCATCCAGATTCTTCTAGGTGTTTGTTTGTGTACCACTTCCCTTATTAGGCTGCTTCTGTCAGTTCCTGAGGGTAGGAAGTGAGTTCCAGGTCTGACTGCTTCAGGTCTAGCCTATGGCTATGCTCAGAGAATGCTGGTGCGGGCCTGAGTGGAAGGCAGTCTGTCCTCACTGCCTCCCTTCTCTCTCACCCAGATGAAAACCCAGTATGAGCAGACCCTGGCCGAGCTAAATCGCTACACCCC .........................................................................................................(((..(((.(((((...))))).)))..)))......((((.(((...(((.((.(((((((........))))))))).))).))).))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGT................................................................................... | 23 | 1 | 5.00 | 5.00 | - | - | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | 1.00 | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGa................................................................................... | 23 | a | 3.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGTt.................................................................................. | 24 | t | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAG.................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................GCGGGCCTGAGTGGAAGGCAGT................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAAGGCAG.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| TGCACTTCATCCAGATTCTTCTAGGTGTTTGTTTGTGTACCACTTCCCTTATTAGGCTGCTTCTGTCAGTTCCTGAGGGTAGGAAGTGAGTTCCAGGTCTGACTGCTTCAGGTCTAGCCTATGGCTATGCTCAGAGAATGCTGGTGCGGGCCTGAGTGGAAGGCAGTCTGTCCTCACTGCCTCCCTTCTCTCTCACCCAGATGAAAACCCAGTATGAGCAGACCCTGGCCGAGCTAAATCGCTACACCCC .........................................................................................................(((..(((.(((((...))))).)))..)))......((((.(((...(((.((.(((((((........))))))))).))).))).))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................AGGCTGCTTCTGTCAGTTCCTGAGGGTA......................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GTCTAGCCTATGGCTATGCTCAGAGAA................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .................................TACCACTTCCCTTAttcg....................................................................................................................................................................................................... | 18 | ttcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................CTTCTCTCTCACCCAcaga................................................... | 19 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................CCAGGTCTGACTGCTTCAGGTCTAGCCTA................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................TTGTTTGTGTACCACTTCCCTTATTA.................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................GGCTGCTTCTGTCAGTTCCTGAGGGTA......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |