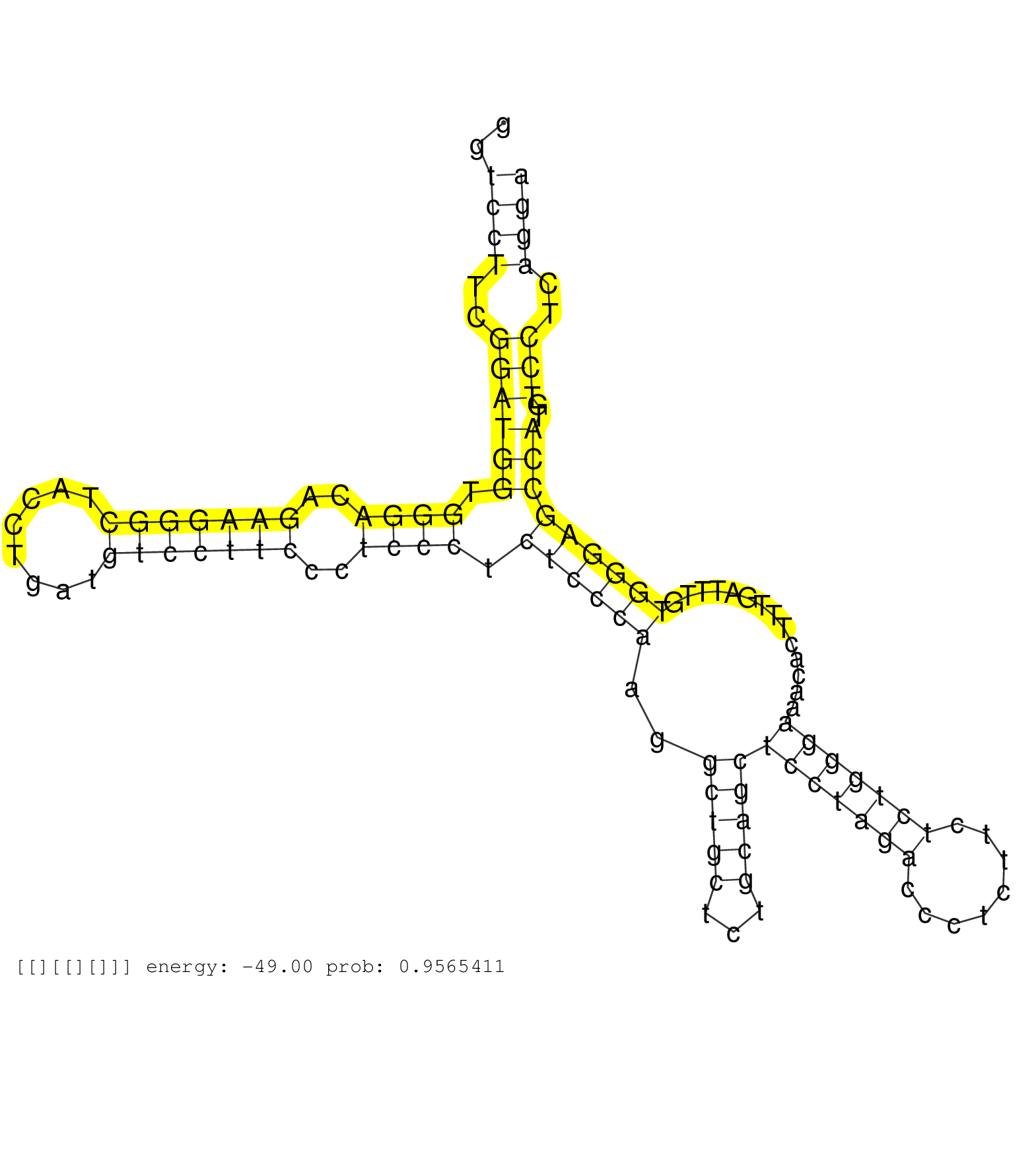

| Gene: P2rx3 | ID: uc008kjr.1_intron_10_0_chr2_84865149_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| GGGATCATCAACCGAGCCGTTCAGCTGCTGATTATCTCCTACTTTGTGGGGTGAGTCCATGGTCCTTCGGATGGTGGGACAGAAGGGCTACCTGATGTCCTTCCCTCCCTCTCCCAAGGCTGCTCTGCAGCTCCTAGACCCTCTTCTCTGGGAAACACTTTGATTTGTGGGAGCCATGTCCTCAGGATTGGCCTCTGCCACTCCCATCTAGACCCGTCTCAGGTGCATTATTTTACCCAGAATCTCTTCC ..............................................................((((..((((((.((((..(((((((........)))))))..)))).((((((..(((((...)))))(((((((........)))))))..............)))))))))..)))..))))............................................................... ............................................................61............................................................................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................TTTGATTTGTGGGAGCCATGTCCTC................................................................... | 25 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGGGAAACACTTTGATTTGTGGGAGCC........................................................................... | 27 | 1 | 4.00 | 4.00 | - | - | - | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................TGGGAAACACTTTGATTTGTGGGAGC............................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTCTCTGGGAAACACTTTGATTTGTGGG............................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TGTGGGGTGAGTCCATGGTCCTTCGGtt.................................................................................................................................................................................. | 28 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TTTGATTTGTGGGAGCCATGTCCTCA.................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................TGATTTGTGGGAGCCATGTCCTCAGG................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TCTGGGAAACACTTTGATTTGTGGGAGC............................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................TTCGGATGGTGGGACAGAAGGGCTACCT............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CACTTTGATTTGTGGGAGCCATGTCCTCAGGA............................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................TCTGGGAAACACTTTGATTTGTGGGA.............................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TTCTCTGGGAAACACTTTGATTTGTGGGAGa............................................................................ | 31 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................................................TGGGAGCCATGTCCTCAta................................................................ | 19 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................CCTGATGTCCTTCCCTCCCTCTCCCA...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................TGGGAAACACTTTGATTTGTGGGAGCCA.......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| GGGATCATCAACCGAGCCGTTCAGCTGCTGATTATCTCCTACTTTGTGGGGTGAGTCCATGGTCCTTCGGATGGTGGGACAGAAGGGCTACCTGATGTCCTTCCCTCCCTCTCCCAAGGCTGCTCTGCAGCTCCTAGACCCTCTTCTCTGGGAAACACTTTGATTTGTGGGAGCCATGTCCTCAGGATTGGCCTCTGCCACTCCCATCTAGACCCGTCTCAGGTGCATTATTTTACCCAGAATCTCTTCC ..............................................................((((..((((((.((((..(((((((........)))))))..)))).((((((..(((((...)))))(((((((........)))))))..............)))))))))..)))..))))............................................................... ............................................................61............................................................................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|