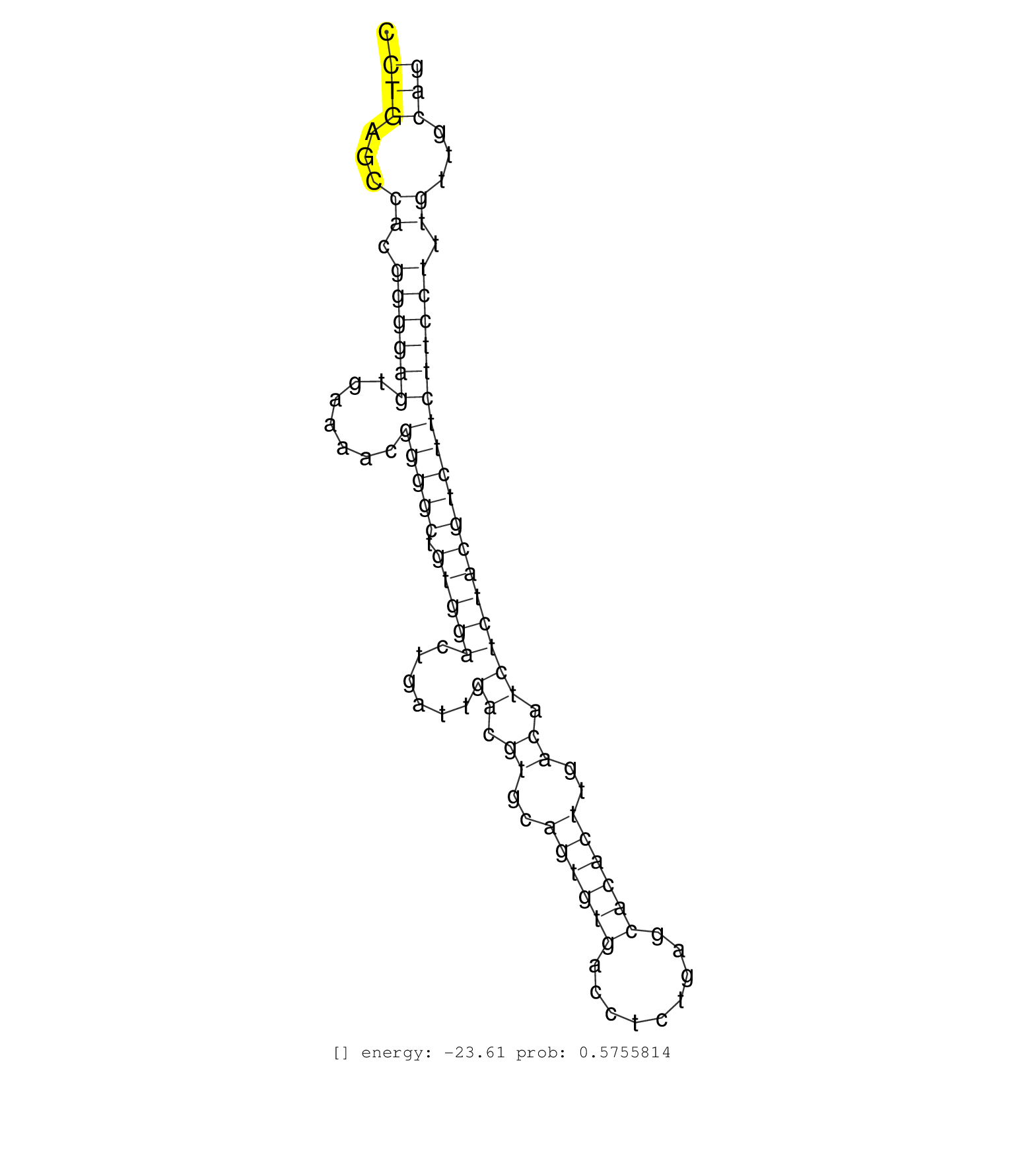

| Gene: Ssfa2 | ID: uc008kgu.1_intron_5_0_chr2_79482347_f.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
| CACTAGTATCATGAGAAAATGTGCCTGTATATCCTTTCTAAGGACTGTGTATCTCATGCATGCGAAGTATCGGAAGATACAGAAAGATATGATGTGAAGACCTGAGCCACGGGGAGTGAAAACGGGGCTGTGGACTGATTGACGTGCAGTGTGACCTCTGAGCACACTTGACATCTCTACGTCTTCTTCCTTTGTTGCAGTGTCTCAGAACTGCTGGAGCTTTACGAGGAAGATCCTGAAGAAATTCTAT .....................................................................................................(((...((.((((((.......(((((.(((((......((.((..((((((.........))))))..)).)))))))))))))))))).))...))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................AGATATGATGTGAAGACCTGAGC............................................................................................................................................... | 23 | 1 | 34.00 | 34.00 | 11.00 | 2.00 | 5.00 | 6.00 | 2.00 | 3.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ....................................................................................AGATATGATGTGAAGACCTGA................................................................................................................................................. | 21 | 1 | 6.00 | 6.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................AAGATATGATGTGAAGACCTGAGC............................................................................................................................................... | 24 | 1 | 6.00 | 6.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ....................................................................................AGATATGATGTGAAGACCTGAG................................................................................................................................................ | 22 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........CATGAGAAAATGTGCCTGTATATC......................................................................................................................................................................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAACTGCTGGAGCTTTACGAGGAAGATC............... | 28 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GTCTCAGAACTGCTGGAGC.............................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................AGATATGATGTGAAGACCT................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAGAACTGCTGGAGCTTTACGA....................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................AGAACTGCTGGAGCTTTACGAGGAAGATC............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................................................................................GTGTCTCAGAACTGCTGGAGCTTTACGAGGAt................... | 32 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GGGGAGTGAAAACGGGGCTGTGGACTGATTGAC........................................................................................................... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAACTGCTGGAGCTTTACGAGGAAGA................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................GTGTCTCAGAACTGCTGGAGCTTTACG........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAACTGCTGGAGCTTTACGAGGAAG.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAACTGCTGGAGCTTTACGAGGAAGAT................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CAGAACTGCTGGAGCTTTACGAGGAA................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................CAGAACTGCTGGAGC.............................. | 15 | 10 | 0.20 | 0.20 | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACTAGTATCATGAGAAAATGTGCCTGTATATCCTTTCTAAGGACTGTGTATCTCATGCATGCGAAGTATCGGAAGATACAGAAAGATATGATGTGAAGACCTGAGCCACGGGGAGTGAAAACGGGGCTGTGGACTGATTGACGTGCAGTGTGACCTCTGAGCACACTTGACATCTCTACGTCTTCTTCCTTTGTTGCAGTGTCTCAGAACTGCTGGAGCTTTACGAGGAAGATCCTGAAGAAATTCTAT .....................................................................................................(((...((.((((((.......(((((.(((((......((.((..((((((.........))))))..)).)))))))))))))))))).))...))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................AGTGTCTCAGAACTccgt...................................... | 18 | ccgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |