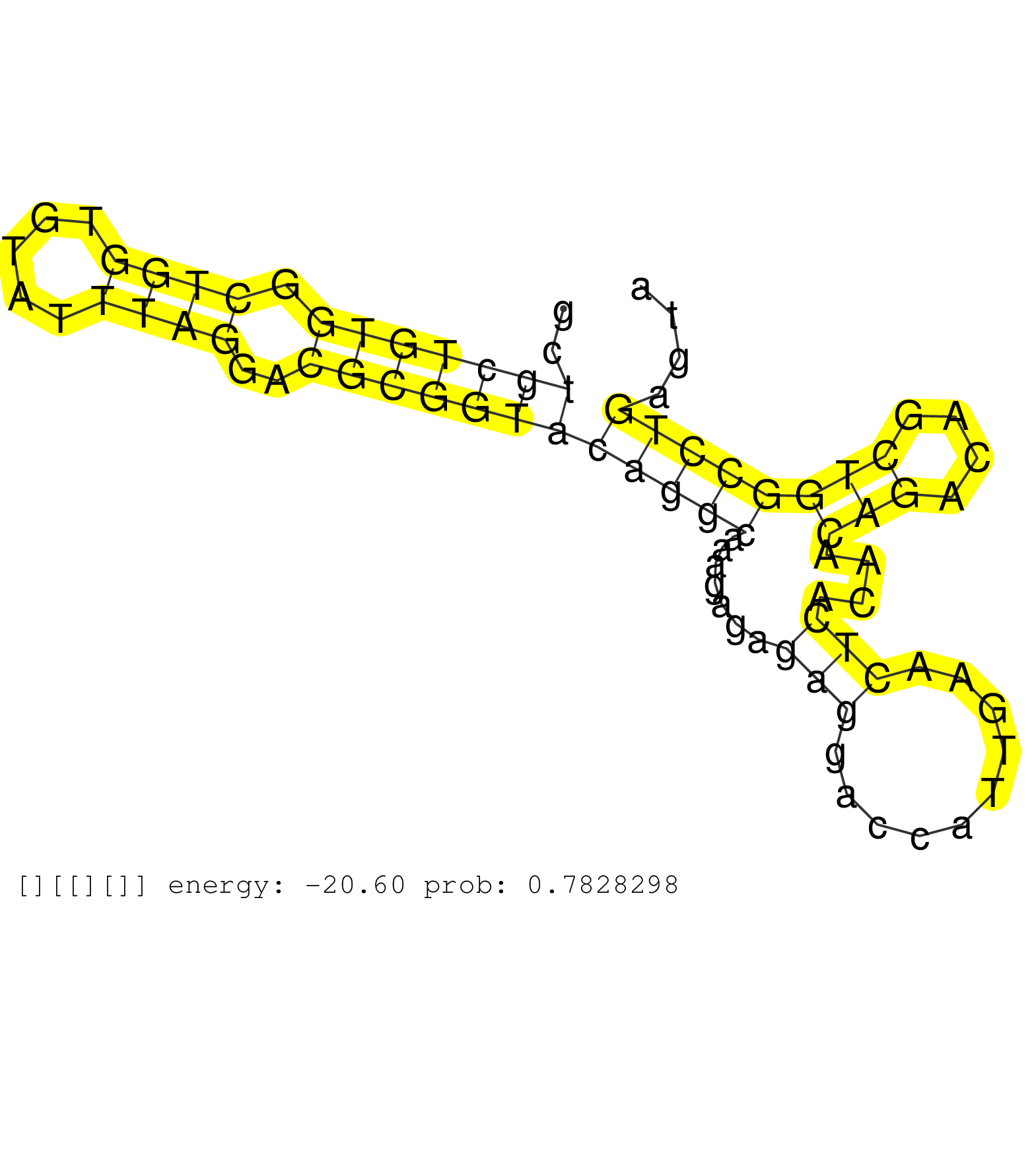
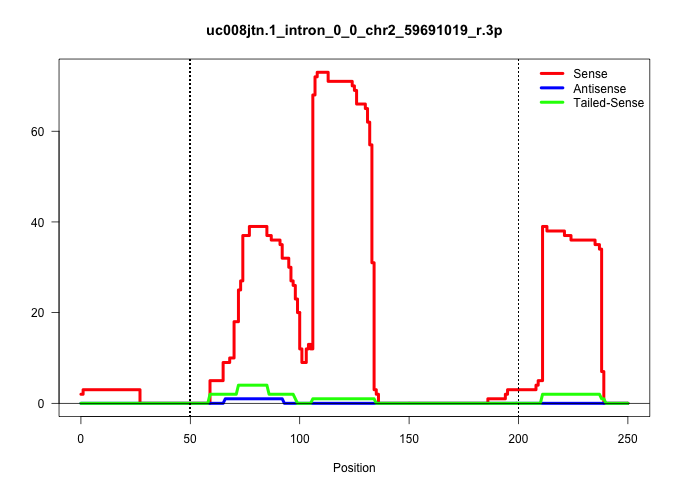
| Gene: Wdsub1 | ID: uc008jtn.1_intron_0_0_chr2_59691019_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| TAGCACTTCACAGAGCACCGTCCGGGCTCAGCGTGCCTTCAGGGCTACGGGCCAGCTGCTGTGGCTGGTGTATTTAGGACGCGGTACAGGCAAAGAGAGAGGACCATTGAACTCACAACAGACAGCTGGCCTGAGTACAGTTCTGCTCACTCACCCTGATCCCCGCTCTTCACCTGACTGAAGCCATGTGCTTTTTGCAGATGGCTACTCCTACGAGAGAGAAGCAATGGAAAGCTGGATCCACAAGAAG ........................................................(((((((.((((.....))))..)))))))(((((.......(((..........)))....(((....))))))))..................................................................................................................... ......................................................55................................................................................137............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................TACGAGAGAGAAGCAATGGAAAGCTGG............ | 27 | 1 | 27.00 | 27.00 | 7.00 | 7.00 | 1.00 | - | 2.00 | 5.00 | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 |
| ..........................................................................................................TTGAACTCACAACAGACAGCTGGCCTG..................................................................................................................... | 27 | 1 | 26.00 | 26.00 | 11.00 | 9.00 | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGAACTCACAACAGACAGCTGGCCTGA.................................................................................................................... | 28 | 1 | 24.00 | 24.00 | 5.00 | 7.00 | 5.00 | 5.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGGACGCGGTACAGGCAAAGAGAGA...................................................................................................................................................... | 26 | 1 | 8.00 | 8.00 | 2.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACGAGAGAGAAGCAATGGAAAGCTGGA........... | 28 | 1 | 8.00 | 8.00 | 2.00 | - | - | - | 1.00 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................TTTAGGACGCGGTACAGGCAAAGAGA........................................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGAACTCACAACAGACAGCTGGCCT...................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TATTTAGGACGCGGTACAGGCAAAGA.......................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGAACTCACAACAGACAGCTGGCCTGA.................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGTGTATTTAGGACGCGGTACAGGCA.............................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGGACGCGGTACAGGCAAAGAGAGAG..................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................TGTGGCTGGTGTATTTAGGACGCGGT..................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CAGGCAAAGAGAGAGGACCATTGAACT......................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GAGGACCATTGAACTCACAACAGACAGC............................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTGGCTGGTGTATTTAGGACGCGGga.................................................................................................................................................................... | 27 | ga | 2.00 | 0.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTGGCTGGTGTATTTAGGACGCGGTA.................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CCATTGAACTCACAACAGACAGCTGGCC....................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TATTTAGGACGCGGTACAGGCAAAGAG......................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................GACGCGGTACAGGCAAAGAGAGAGGACC................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGTGTATTTAGGACGCGGTACAGGC............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGAACTCACAACAGACAGCTGGCCTGAGT.................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGAGGACCATTGAACTCACAACAGACAGC............................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TATTTAGGACGCGGTACAGGCAAAG........................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACGAGAGAGAAGCAATGGAAAGCTGt............ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................TTAGGACGCGGTACAGGCAAAGAGAG....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GAGAGGACCATTGAACTtgc...................................................................................................................................... | 20 | tgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................AGAGGACCATTGAACTCACAACAGACAG............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................TTGCAGATGGCTACTCCTACGAGAGAG............................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTGAACTCACAACAGACAGCTGGCCTGAt................................................................................................................... | 29 | t | 1.00 | 24.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTGGCTGGTGTATTTAGGACGCGGTAC................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACGAGAGAGAAGCAATGGAAAGCTGGAa.......... | 29 | a | 1.00 | 8.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TGTATTTAGGACGCGGTACAGGCAAAG........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CCATTGAACTCACAACAGACAGCTGGC........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................CCTACGAGAGAGAAGCAATGGAAAGC............... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTTAGGACGCGGTACAGGCAAAGAGt........................................................................................................................................................ | 26 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GAACTCACAACAGACAGCTGGCCTGA.................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................TTGAACTCACAACAGACAGCTGGCCTGAG................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TGTGCTTTTTGCAGATGGCTACTCCTA..................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGATGGCTACTCCTACGAGAGAGAAG.......................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................TATTTAGGACGCGGTACAGGCAAAGAGAG....................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCCTACGAGAGAGAAGCAATGGAAAGCTG............. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGAACTCACAACAGACAGCTGGCCTGAGT.................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTTAGGACGCGGTACAGGCAAAGAGAt....................................................................................................................................................... | 27 | t | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATTGAACTCACAACAGACAGCTGGCCT...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTTAGGACGCGGTACAGGCAAAGAGAG....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................GAGAGGACCATTGAACTCACAACAGACA.............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .AGCACTTCACAGAGCACCGTCCGGGC............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACGAGAGAGAAGCAATGGAAAGCcgg............ | 27 | cgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................CATTGAACTCACAACAGACAGCTGGCC....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTTAGGACGCGGTACAGGCAAAGAG......................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................TTAGGACGCGGTACAGGCAAAGAGAGAG..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TAGCACTTCACAGAGCACCGTCCGGGCTCAGCGTGCCTTCAGGGCTACGGGCCAGCTGCTGTGGCTGGTGTATTTAGGACGCGGTACAGGCAAAGAGAGAGGACCATTGAACTCACAACAGACAGCTGGCCTGAGTACAGTTCTGCTCACTCACCCTGATCCCCGCTCTTCACCTGACTGAAGCCATGTGCTTTTTGCAGATGGCTACTCCTACGAGAGAGAAGCAATGGAAAGCTGGATCCACAAGAAG ........................................................(((((((.((((.....))))..)))))))(((((.......(((..........)))....(((....))))))))..................................................................................................................... ......................................................55................................................................................137............................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................GGTGTATTTAGGACGCGGTACAGGCAA............................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |