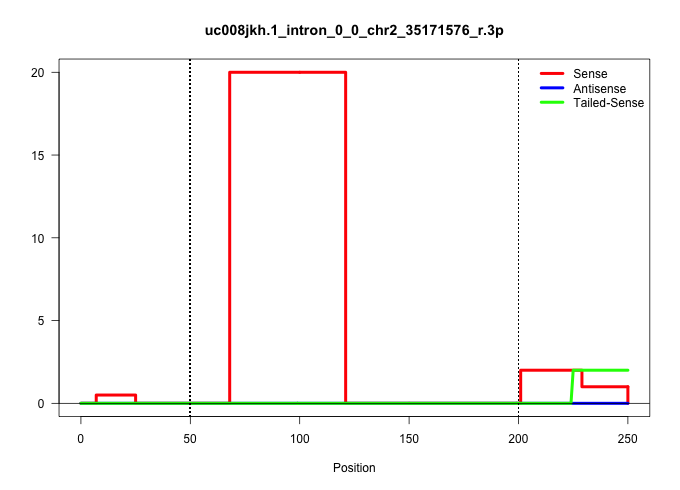
| Gene: Stom | ID: uc008jkh.1_intron_0_0_chr2_35171576_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(2) PIWI.ip |
(8) TESTES |
| TGGGAGACAAGGCAGCAGTAGAAGAGAAAGCCTAGAGCCAGACACATCCTGACACAATTCAGTTTCAGGAAGAAGAGCCCCTGGGATCTGAGTGTCTAGGGATTGATGGGCTCCTGGCAAGTCCCGTATGCTGTCTTACTCGAAGACCACTATGTGGCACCATGCTATTTGCCAGCTGATCTCTTCTTCCCCCTGTGCAGGTGATTGCAGCTGAAGGGGAAATGAATGCGTCCAGAGCTCTGAAAGAAGC ....................................................................................................((..(((.(((...((((((((...(((((.((((....................)))).))))).))))))))))).)))...))................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................GAAGAAGAGCCCCTGGGATCTGAGTGTCTAGGGATTGATGGGCTCCTGGCAA.................................................................................................................................. | 52 | 1 | 20.00 | 20.00 | 20.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGAATGCGTCCAGAGCTCTGAAAGA... | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................................ATGCGTCCAGAGCTCTGAAAGAAGCgtct | 29 | gtct | 2.00 | 0.00 | - | 1.00 | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................TGATTGCAGCTGAAGGGGAAATGAAT....................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................................TGATTGCAGCTGAAGGGGAAATGAATGC..................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GCGTCCAGAGCTCTGAAAGAAGC | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................................................................TGAATGCGTCCAGAGCTCTGAAAG.... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| .......CAAGGCAGCAGTAGAAGA................................................................................................................................................................................................................................. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 |
| TGGGAGACAAGGCAGCAGTAGAAGAGAAAGCCTAGAGCCAGACACATCCTGACACAATTCAGTTTCAGGAAGAAGAGCCCCTGGGATCTGAGTGTCTAGGGATTGATGGGCTCCTGGCAAGTCCCGTATGCTGTCTTACTCGAAGACCACTATGTGGCACCATGCTATTTGCCAGCTGATCTCTTCTTCCCCCTGTGCAGGTGATTGCAGCTGAAGGGGAAATGAATGCGTCCAGAGCTCTGAAAGAAGC ....................................................................................................((..(((.(((...((((((((...(((((.((((....................)))).))))).))))))))))).)))...))................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) |
|---|