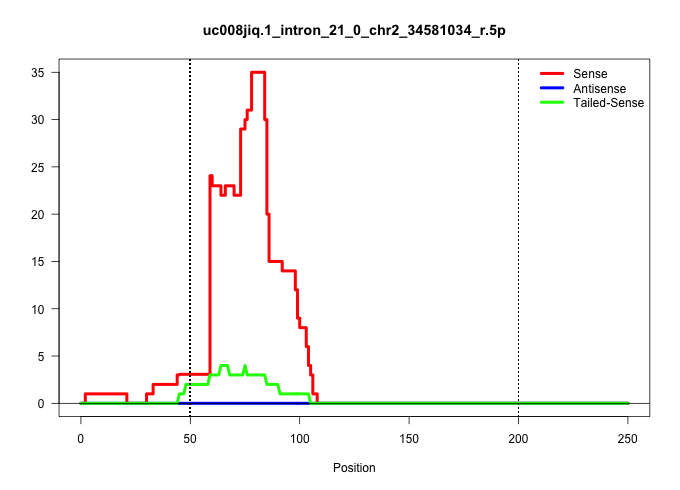
| Gene: Gapvd1 | ID: uc008jiq.1_intron_21_0_chr2_34581034_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(19) TESTES |
| CTGAAGAAGGAGATCCCCGGACCAAAAATAGCCTTGGAAAATTTGACAAAGTAGGAATGAGTGTTGTGTCATGAGAAATACGGTTCGTAGACTATAAGATGTCCAGGAGTGCTAACCCACACCTGTAATCTCAGTTGAGGTGGGAGGATTGCCTTGAGTCCAAGGTTAGCCTAGATAACAGTGAGTCCTCCTTACAAACAAAGGAAACATACTGATTTGGTGCACAACTTTTACTCTGCATTCACTGTGA ............................................................((.((((((((((((........))))).))).)))).))...................................................................................................................................................... ..................................................51..................................................103................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................AGTGTTGTGTCATGAGAAATACGGTT..................................................................................................................................................................... | 26 | 1 | 10.00 | 10.00 | 1.00 | 1.00 | 2.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...........................................................AGTGTTGTGTCATGAGAAATACGGTTC.................................................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | 2.00 | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGTGTTGTGTCATGAGAAATACGGT...................................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................AGAAATACGGTTCGTAGACTATAAGA....................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................AGAAATACGGTTCGTAGACTATAAG........................................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TACGGTTCGTAGACTATAAGATGTCC.................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AAATACGGTTCGTAGACTATAAGATGgcca................................................................................................................................................. | 30 | gcca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................GCCTTGGAAAATTTGACAAAGTAGGAATGA.............................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TACGGTTCGTAGACTATAAGATGTCCAG................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GAAGAAGGAGATCCCCGGA..................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGTGTTGTGTCATGAGAAATACGGgc..................................................................................................................................................................... | 26 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................TGTGTCATGAGAAATACGGTTCGTAGt............................................................................................................................................................... | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TACGGTTCGTAGACTATAAGATGTCCA................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................AATACGGTTCGTAGACTATAAGATGTCCAG................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................GTAGACTATAAGATGTCCAGGA.............................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................GACAAAGTAGGAATGAGTGTTGTGTC.................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................AAATACGGTTCGTAGACTATAAGATGTC................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TGTCATGAGAAATACGGTTCGTAGAC.............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................GAAATACGGTTCGTAGACTATAAGATGTCC.................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................AGAAATACGGTTCGTAGACTATAAGAT...................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................ACAAAGTAGGAATGAGTGTTGTGTCATGAGt.............................................................................................................................................................................. | 31 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................AGAAATACGGTTCGTAGACTATAAGATGTC................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AAGTAGGAATGAGTGTTtaa...................................................................................................................................................................................... | 20 | taa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TTGGAAAATTTGACAAAGTAGGAATGAGTGT.......................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................ACAAAGTAGGAATGA.............................................................................................................................................................................................. | 15 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |
| CTGAAGAAGGAGATCCCCGGACCAAAAATAGCCTTGGAAAATTTGACAAAGTAGGAATGAGTGTTGTGTCATGAGAAATACGGTTCGTAGACTATAAGATGTCCAGGAGTGCTAACCCACACCTGTAATCTCAGTTGAGGTGGGAGGATTGCCTTGAGTCCAAGGTTAGCCTAGATAACAGTGAGTCCTCCTTACAAACAAAGGAAACATACTGATTTGGTGCACAACTTTTACTCTGCATTCACTGTGA ............................................................((.((((((((((((........))))).))).)))).))...................................................................................................................................................... ..................................................51..................................................103................................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................GGAGTGCTAACCgga..................................................................................................................................... | 15 | gga | 4.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ................ACCAAAAATAGCCtggc......................................................................................................................................................................................................................... | 17 | tggc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................CCCACACCTGTAATCTCAgc..................................................................................................................... | 20 | gc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |