
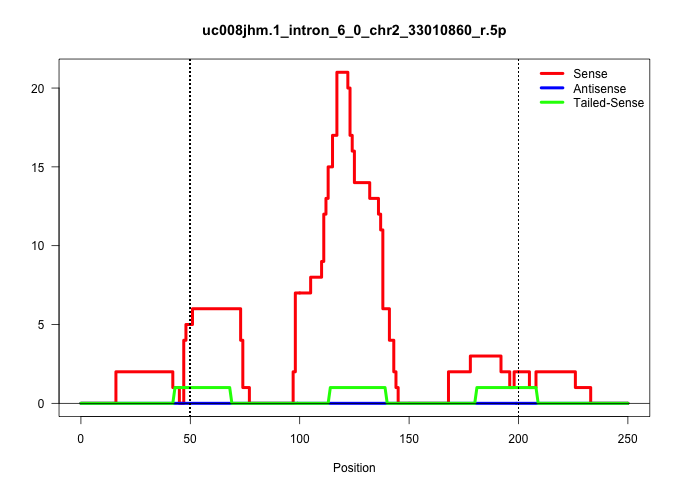
| Gene: Ralgps1 | ID: uc008jhm.1_intron_6_0_chr2_33010860_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(13) TESTES |
| GGGCCTCACCCACACCTCCTCCACCGCCATCACCAACGGACTCTCCCTAGGTAAGCGTCTCTGGCCTGTGCATACAGGCAGGGGCAGCAGGCCCAGTTTGTGCTCTGTGTCTGTCTCTGTGTCGCTGTACTGTGCCCAGTGGCCCCTCACCCTTAAAGAATGGCTCCCTACAAGAGTCTTCTGCTGCAGGAAGAGCCCTGGCTTGGCCTTCTCCTTCTGGTGGTCCCCAAATGGACAGTCAGATTGAAGA ........................................................(((.((((...(((((((((((((((((((((((((.((...)).)).)))...))))))))))....))))).))))))))).)))........................................................................................................... ..................................................51............................................................................................145....................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................TCTGTGTCGCTGTACTGTGCCCAGTG............................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................TGTGTCGCTGTACTGTGCCCAGTGGC........................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGTCTCTGTGTCGCTGTACTGTGCCCA................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - |
| ..................................................................................................TGTGCTCTGTGTCTGTCTCTGTGTC............................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGTGCTCTGTGTCTGTCTCTGTGTCGC............................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ...............................................TAGGTAAGCGTCTCTGGCCTGTGCAT................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .................................................................................................................TCTCTGTGTCGCTGTACTGTGCCCA................................................................................................................ | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................GCTCTGTGTCTGTCTCTGTGTCGCT............................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - |
| ...............................................TAGGTAAGCGTCTCTGGCCTGTGCATA................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ................................................AGGTAAGCGTCTCTGGCCTGTGCATA................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGTGTCTGTCTCTGTGTCGCTGTACTG...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CTCTGTGTCGCTGTACTGTGCCCAGc.............................................................................................................. | 26 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TACAAGAGTCTTCTGCTGCAGGAAGAGC...................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................TCCTCCACCGCCATCACCAACGGACT................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................TGGCTTGGCCTTCTCCTTCTGGTGGTCC........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TAAGCGTCTCTGGCCTGTGCATACAG............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGTGCTCTGTGTCTGTCTCTGTGT................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTGTCGCTGTACTGTGCCCAGTGGCC.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TTGTGCTCTGTGTCTGTCTCTGTGTC............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................TCCCTAGGTAAGCGTCTCTGGCCcgt..................................................................................................................................................................................... | 26 | cgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TGGACAGTCAGATTGAAGA | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................GTCTCTGTGTCGCTGTACTGTGCCCA................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TACAAGAGTCTTCTGCTGCAGGAA.......................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................TTGTGCTCTGTGTCTGTCTCTGTGTCG.............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................TGCTGCAGGAAGAGCCCTGGCTTGGCCa......................................... | 28 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................TCCTCCACCGCCATCACCAACGGACTCTC............................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TTCTCCTTCTGGTGGTCCCCAAATG................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGTCTCTGTGTCGCTGTACTGTGCCC................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTCTGCTGCAGGAAGAGCCCTGGCTTG............................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGTCTCTGTGTCGCTGTACTGTGCC.................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TGTGTCGCTGTACTGTGCCCAGTGGCCC......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GGGCCTCACCCACACCTCCTCCACCGCCATCACCAACGGACTCTCCCTAGGTAAGCGTCTCTGGCCTGTGCATACAGGCAGGGGCAGCAGGCCCAGTTTGTGCTCTGTGTCTGTCTCTGTGTCGCTGTACTGTGCCCAGTGGCCCCTCACCCTTAAAGAATGGCTCCCTACAAGAGTCTTCTGCTGCAGGAAGAGCCCTGGCTTGGCCTTCTCCTTCTGGTGGTCCCCAAATGGACAGTCAGATTGAAGA ........................................................(((.((((...(((((((((((((((((((((((((.((...)).)).)))...))))))))))....))))).))))))))).)))........................................................................................................... ..................................................51............................................................................................145....................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................CTGGCTTGGCCTTCTCctca..................................... | 20 | ctca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |