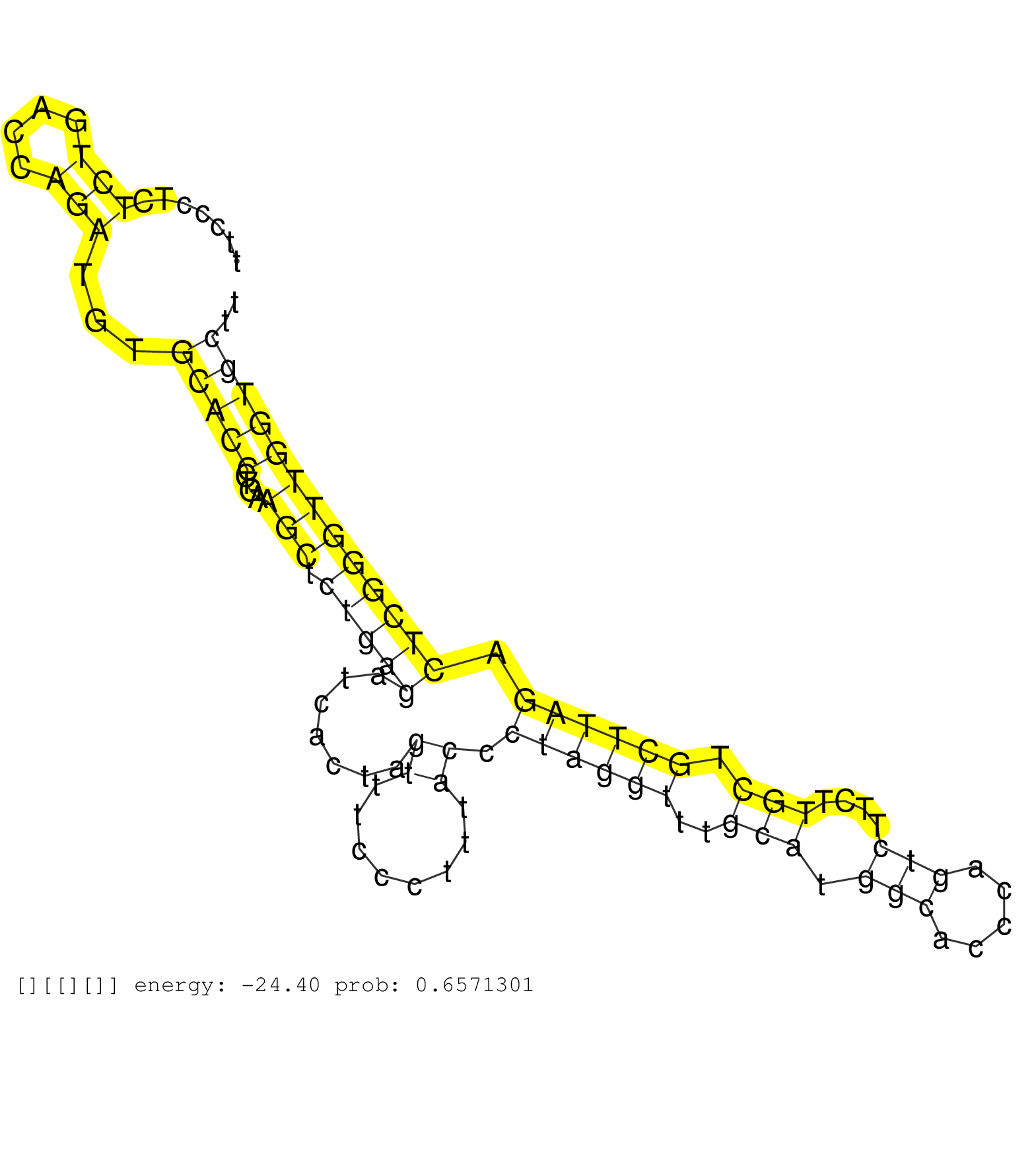
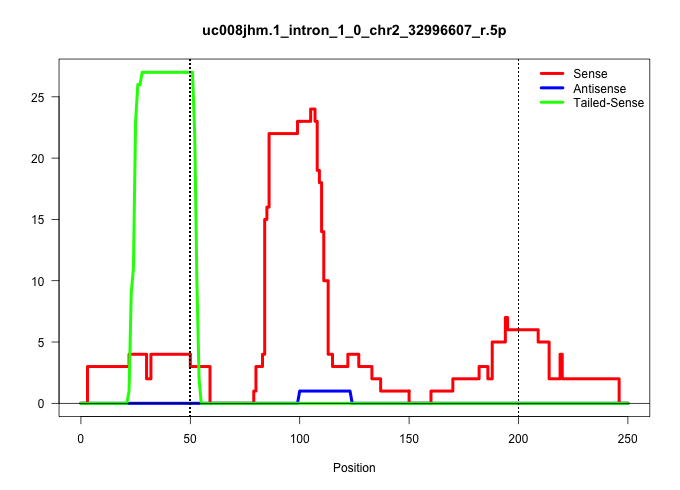
| Gene: Ralgps1 | ID: uc008jhm.1_intron_1_0_chr2_32996607_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(18) TESTES |
| AGATGACCCCGAGCACCCAGATATCTTCCAGCTGAATAACCCTGACAAAGGTGGGTGGCAGCCTGGGCTGGAGCCACTGTTCCCTCTCTGACCAGATGTGCACCCTGAAGCTCTGAGATCACTAGTTTCCCTTTACCCTAGGTTTGCATGGCACCCAGTCTTCTTGCTGCTTAGACTCGGGTTGGTGCTTCTGTTGAGCAGTGAGAGAGGCCTTGCCTTTGACTTGATGGGTCTGTCCTTCTTCGGTCTG ......................................................................................(((....)))...(((((....(((.(((((.......((........)).((((((..(((.(((.....)))....))).)))))).))))))))))))).............................................................. ...............................................................................80............................................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................TTCCAGCTGAATAACCCTGACAAAGGca..................................................................................................................................................................................................... | 28 | ca | 6.00 | 0.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTCTGACCAGATGTGCACCCTGAAGC........................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TCTGACCAGATGTGCACCCTGAAGCTC......................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................TTCCAGCTGAATAACCCTGACAAAGGcaa.................................................................................................................................................................................................... | 29 | caa | 3.00 | 0.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTTCCAGCTGAATAACCCTGACAAAGGca..................................................................................................................................................................................................... | 30 | ca | 3.00 | 0.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TTCTGTTGAGCAGTGAGAGAGGCCTT.................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTCTGACCAGATGTGCACCCTGAAG............................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTAGACTCGGGTTGGTGCTTCTGTT....................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..........................TCCAGCTGAATAACCCTGACAAAGGcaa.................................................................................................................................................................................................... | 28 | caa | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................TGAATAACCCTGACAAAGGTGGGTGGC............................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGACCCCGAGCACCCAGATATCTTCCA............................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TCTCTGACCAGATGTGCACCCTGA.............................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGACTTGATGGGTCTGTCCTTCTTCGG.... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CTTCCAGCTGAATAACCCTGACAAAGGca..................................................................................................................................................................................................... | 29 | ca | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .........................TTCCAGCTGAATAACCCTGACAAAGGc...................................................................................................................................................................................................... | 27 | c | 2.00 | 0.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTTCCAGCTGAATAACCCTGACAAAGGcat.................................................................................................................................................................................................... | 31 | cat | 2.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TGAGCAGTGAGAGAGGCCTTGCCTTT.............................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................CAGCTGAATAACCCTGACAAAGGcaat................................................................................................................................................................................................... | 27 | caat | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTTCCAGCTGAATAACCCTGACAAAGGcatt................................................................................................................................................................................................... | 32 | catt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................TCTTCCAGCTGAATAACCCTGACAAAGGc...................................................................................................................................................................................................... | 29 | c | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGACCCCGAGCACCCAGATATCTTCCAGCT......................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ATCTTCCAGCTGAATAACCCTGACAAAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................TCCAGCTGAATAACCCTGACAAAGGca..................................................................................................................................................................................................... | 27 | ca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TCCCTCTCTGACCAGATGTGCACCCTGA.............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGTTTCCCTTTACCCTAGGTTTGCATG.................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ATCTTCCAGCTGAATAACCCTGACAAAGGc...................................................................................................................................................................................................... | 30 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................GAATAACCCTGACAAAGGTGGGTGGC............................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCTTCCAGCTGAATAACCCTGACAAAGGa...................................................................................................................................................................................................... | 29 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGGTGCTTCTGTTGAGCAGTGAGAGAG......................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CACTGTTCCCTCTCTGACCAGATGTGCAC................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................TCTCTGACCAGATGTGCACCCTGAAGCTC......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CTCTGACCAGATGTGCACCCTGAAGCTC......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGAAGCTCTGAGATCACTAGTTTCCCTT..................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCTGAGATCACTAGTTTCCCTTTACC................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................TTCCAGCTGAATAACCCTGACAAAGGcat.................................................................................................................................................................................................... | 29 | cat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TCTGACCAGATGTGCACCCTGA.............................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTCTCTGACCAGATGTGCACCCTGAAG............................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TCTGACCAGATGTGCACCCTGAAGCTCTG....................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TTCCCTCTCTGACCAGATGTGCACCCTG............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TTCTTGCTGCTTAGACTCGGGTTGGT................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GCACCCTGAAGCTCTGAGATCACTAGTT........................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TCCCTCTCTGACCAGATGTGCACCCTGAA............................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGATGACCCCGAGCACCCAGATATCTTCCAGCTGAATAACCCTGACAAAGGTGGGTGGCAGCCTGGGCTGGAGCCACTGTTCCCTCTCTGACCAGATGTGCACCCTGAAGCTCTGAGATCACTAGTTTCCCTTTACCCTAGGTTTGCATGGCACCCAGTCTTCTTGCTGCTTAGACTCGGGTTGGTGCTTCTGTTGAGCAGTGAGAGAGGCCTTGCCTTTGACTTGATGGGTCTGTCCTTCTTCGGTCTG ......................................................................................(((....)))...(((((....(((.(((((.......((........)).((((((..(((.(((.....)))....))).)))))).))))))))))))).............................................................. ...............................................................................80............................................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................GTCCTTCTTCGgtgg..... | 15 | gtgg | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CACCCTGAAGCTCTGAGATCACTA.............................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................AGTTTCCCTTTACCCgcg................................................................................................................ | 18 | gcg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |