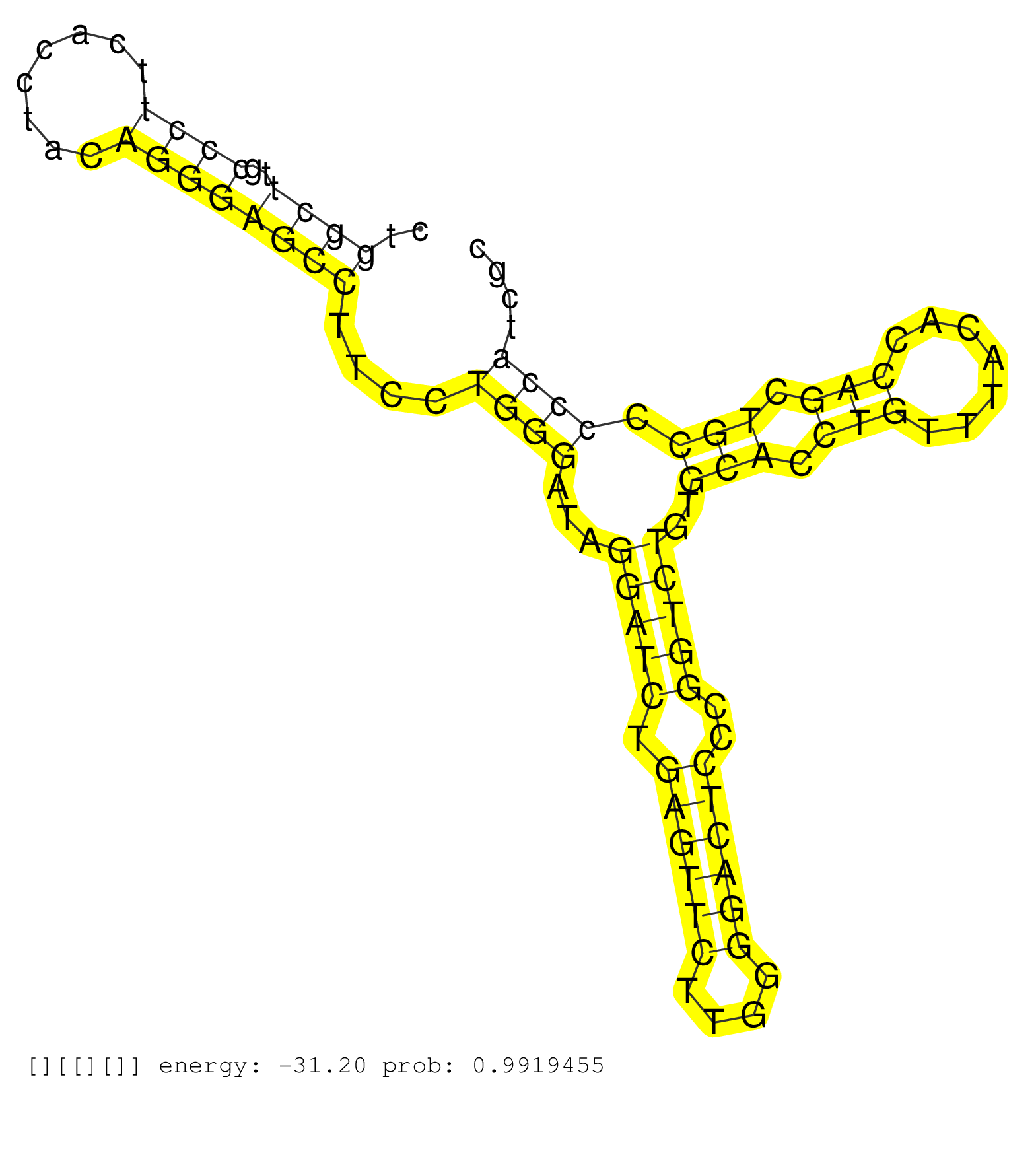
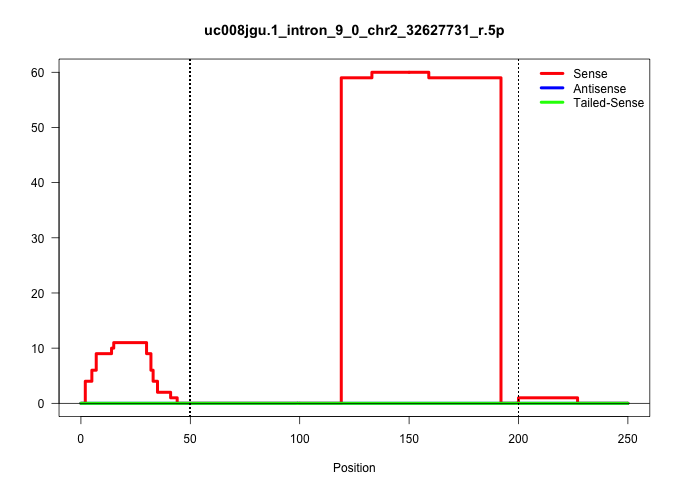
| Gene: Ttc16 | ID: uc008jgu.1_intron_9_0_chr2_32627731_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(4) PIWI.ip |
(11) TESTES |
| CAGGGAATAACAAGTACCTGAACCGGCTGGCCTTTGTGCTCTACCTACAGGTGCCTCAGCTGCCCCACCCGCAGGGGACTTCTCACCTTATCCCCAGCTACTGGCTTGCCCTTCACCTACAGGGAGCCTTCCTGGGATAGGATCTGAGTTCTTGGGGACTCCCGGTCTGTGCACCTGTTTACACCAGCTGCCCCCATCGCTGATTTATTAATTTAGCTGGAGGTGTCTAGAACTTCTGCAGTAAGGCTCT ......................................................................................................((((..((((........))))))))....((((...(((((.((((((....))))))..)))))..(((.(((.......))).))).))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................CAGGGAGCCTTCCTGGGATAGGATCTGAGTTCTTGGGGACTCCCGGTCTGTG............................................................................... | 52 | 1 | 60.00 | 60.00 | 60.00 | - | - | - | - | - | - | - | - | - | - |
| ..GGGAATAACAAGTACCTGAACCGGCTGGCCT......................................................................................................................................................................................................................... | 31 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..GGGAATAACAAGTACCTGAACCGGCTGG............................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .....AATAACAAGTACCTGAACCGGCTGGCC.......................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .......TAACAAGTACCTGAACCGGCTGGCCTTT....................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ...............ACCTGAACCGGCTGGCCTTTGTGCTC................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......TAACAAGTACCTGAACCGGCTGGCC.......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................TGATTTATTAATTTAGCTGGAGGTGTC....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................GGGATAGGATCTGAGTTCTTGGGGAC........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............TACCTGAACCGGCTGGCCTTTGTGCTCTAC.............................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TCCCGGTCTGTGCACC........................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 |
| CAGGGAATAACAAGTACCTGAACCGGCTGGCCTTTGTGCTCTACCTACAGGTGCCTCAGCTGCCCCACCCGCAGGGGACTTCTCACCTTATCCCCAGCTACTGGCTTGCCCTTCACCTACAGGGAGCCTTCCTGGGATAGGATCTGAGTTCTTGGGGACTCCCGGTCTGTGCACCTGTTTACACCAGCTGCCCCCATCGCTGATTTATTAATTTAGCTGGAGGTGTCTAGAACTTCTGCAGTAAGGCTCT ......................................................................................................((((..((((........))))))))....((((...(((((.((((((....))))))..)))))..(((.(((.......))).))).))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................GTGCTCTACCTACAGtgt........................................................................................................................................................................................................ | 18 | tgt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................GTGCTCTACCTACAGtgtt........................................................................................................................................................................................................ | 19 | tgtt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |