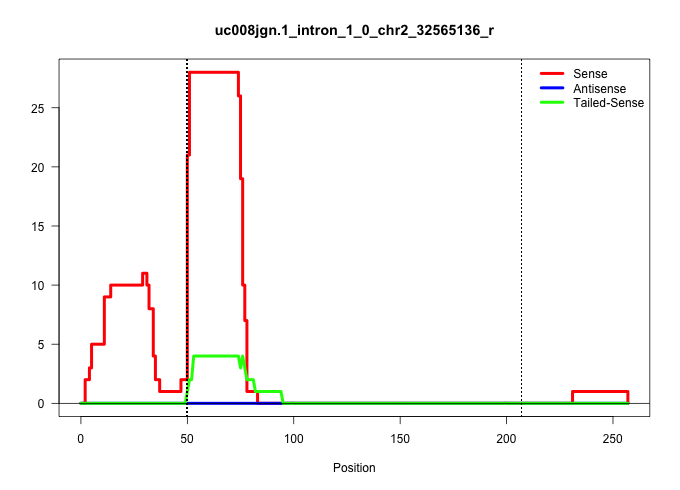
| Gene: Cdk9 | ID: uc008jgn.1_intron_1_0_chr2_32565136_r | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| CACCAACCGTGTGGTGACATTGTGGTACCGGCCTCCGGAGCTGCTGCTCGGTGAGGACTCCCGAGCGGGCAGAGGGGGGCAGGGGCCGGGTGCCTACCTGGCCACAGCCCCCATTGCCTGGGGCCTTTTGAGCCATCAGCTCTGGGACGTGGAGTCTCAGGAGGCCCTTGGGCTTAAGAGGCACTCCCGGTACCCTCTTCTTCTCAGGAGAGCGGGACTACGGCCCGCCCATTGACCTTTGGGGTGCTGGGTGCATC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGGACTCCCGAGCGGGCAGAGGG..................................................................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | 2.00 | - | - | - | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGACTCCCGAGCGGGCAGAGGGGG................................................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGAGGACTCCCGAGCGGGCAGAGG...................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGACTCCCGAGCGGGCAGAGGGG.................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....ACCGTGTGGTGACATTGTGGTACCGGCCT............................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGTGACATTGTGGTACCGGCCTC.............................................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - |
| ...................................................TGAGGACTCCCGAGCGGGCAGAGGG..................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TGGTGACATTGTGGTACCGGCCT............................................................................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGACTCCCGAGCGGGCAGAGGGGG................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGACTCCCGAGCGGGCAGAGG...................................................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TCGGTGAGGACTCCCGAGCGGGCAGAGG...................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ....AACCGTGTGGTGACATTGTGGTACCGGC................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TTGACCTTTGGGGTGCTGGGTGCATC | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................GGCCTCCGGAGCTGCTGC.................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................TGAGGACTCCCGAGCGGGCAGAGGGa.................................................................................................................................................................................... | 26 | a | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGACTCCCGAGCGGGCAGAG....................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CCAACCGTGTGGTGACATTGTGGTACCGGC................................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................AGGACTCCCGAGCGGGCAGAGGGGGGCta............................................................................................................................................................................... | 29 | ta | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGACATTGTGGTACCGGCCTCCG............................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGACTCCCGAGCGGGCAGAGt...................................................................................................................................................................................... | 25 | t | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..CCAACCGTGTGGTGACATTGTGGTACCGG.................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGAGGACTCCCGAGCGGGCAGAGGGGGGCAGG.............................................................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGACTCCCGAGCGGGCAGAG....................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGGACTCCCGAGCGGGCAGAGGGtt................................................................................................................................................................................... | 25 | tt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GGGCAGGGGCCGGGTGacg.................................................................................................................................................................. | 19 | acg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| CACCAACCGTGTGGTGACATTGTGGTACCGGCCTCCGGAGCTGCTGCTCGGTGAGGACTCCCGAGCGGGCAGAGGGGGGCAGGGGCCGGGTGCCTACCTGGCCACAGCCCCCATTGCCTGGGGCCTTTTGAGCCATCAGCTCTGGGACGTGGAGTCTCAGGAGGCCCTTGGGCTTAAGAGGCACTCCCGGTACCCTCTTCTTCTCAGGAGAGCGGGACTACGGCCCGCCCATTGACCTTTGGGGTGCTGGGTGCATC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|