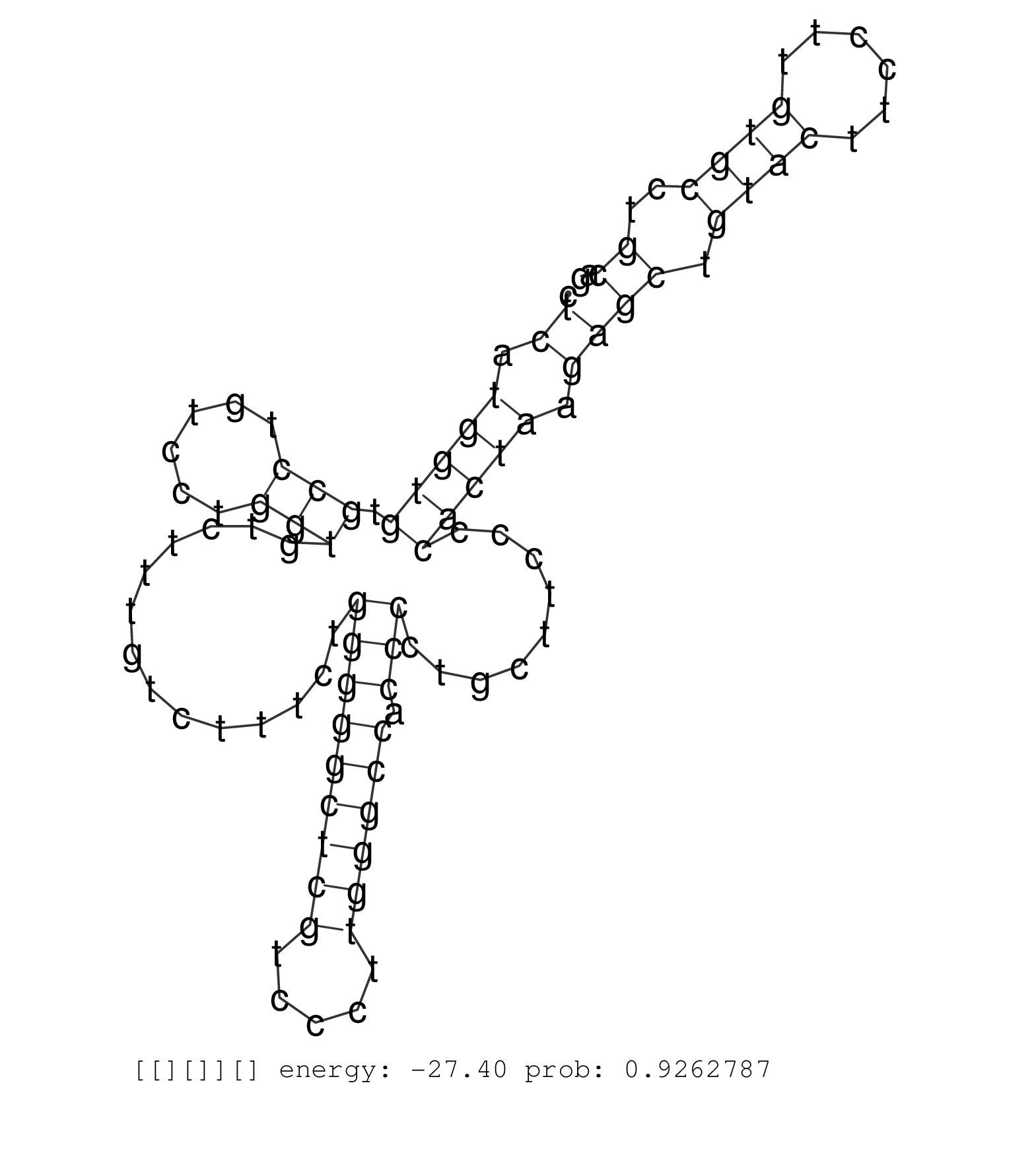
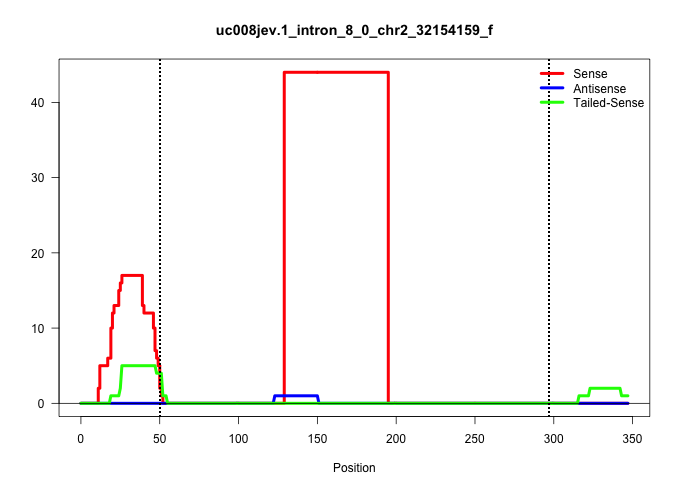
| Gene: Golga2 | ID: uc008jev.1_intron_8_0_chr2_32154159_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(17) TESTES |
| TCTTCCCACAGAAACAACAGAACCAAGACACTCTGGATCAACTGGAGAAAGTAATGTGATAGATTCTATGTTCGCTCTTTTCATGACTGATGGGGGTTGGGGAGACCTCAGAGGTGCAGGCTTCGGTCTTGCTTTCTTCTATCCTGGAGAAGACTCACCCCACAGGGTGTCCTCCATAGGATCTTTGATAACCTGACCTCATGGTGTGCCTGTCCTGGTGTCTTTGTCTTTCTGGGGGCTCGTCCCTTGGGCCACCCCTGCTTCCCCACTAAGAGCTGTACTTCCTTGTGCCTGCAGGAGAAGAAGGATTATCAGCAGAAGCTGGCAAAGGAGCAGGGCTCTCTCAG ......................................................................................................................................................................................................((.(((((.(((......)))..............(((((((((.....)))))).))).........))))).))((.((((......))))..)).................................................... .....................................................................................................................................................................................................198................................................................................................297................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................TGCTTTCTTCTATCCTGGAGAAGACTCACCCCACAGGGTGTCCTCCATAGGA...................................................................................................................................................................... | 52 | 1 | 44.00 | 44.00 | 44.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........AAACAACAGAACCAAGACACTCTGGATC.................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GAACCAAGACACTCTGGATCAACTGGAG............................................................................................................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............AACAACAGAACCAAGACACTCTGGATC.................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GACACTCTGGATCAACTGGAGAAAGa....................................................................................................................................................................................................................................................................................................... | 26 | a | 2.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................ACTAAGAGCTGTACTTCCT............................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................AAGACACTCTGGATCAACTGGAGAAA......................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................ACCAAGACACTCTGGATCAACTGGA............................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AACCAAGACACTCTGGATCAACTGGAGA........................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GAACCAAGACACTCTGGATCAACTGGAaa........................................................................................................................................................................................................................................................................................................... | 29 | aa | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................ATCAACTGGAGAAAGagaa.................................................................................................................................................................................................................................................................................................... | 19 | agaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................GACACTCTGGATCAACTGGAGAAAGagaa.................................................................................................................................................................................................................................................................................................... | 29 | agaa | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................GACACTCTGGATCAACTGGAGAAAG........................................................................................................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................AACCAAGACACTCTGGATCAACTGGAGAAA......................................................................................................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................GAACCAAGACACTCTGGATCAACTGGA............................................................................................................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................AGACACTCTGGATCAACTGGAGAAAGa....................................................................................................................................................................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................CAGAACCAAGACACTCTGGATCAACTGGAG............................................................................................................................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................GGCAAAGGAGCAGGGCTCTCTCAGaaa | 27 | aaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GAACCAAGACACTCTGGATCAACTGGAGAAA......................................................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................AGAAGCTGGCAAAGGAGCAGGGCTCTt.... | 27 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............AACAACAGAACCAAGACACTCTGGATCA................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................AGACACTCTGGATCAACTGGAGAAAGT....................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AAGACACTCTGGATCAACTGGAGAA.......................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TCTTCCCACAGAAACAACAGAACCAAGACACTCTGGATCAACTGGAGAAAGTAATGTGATAGATTCTATGTTCGCTCTTTTCATGACTGATGGGGGTTGGGGAGACCTCAGAGGTGCAGGCTTCGGTCTTGCTTTCTTCTATCCTGGAGAAGACTCACCCCACAGGGTGTCCTCCATAGGATCTTTGATAACCTGACCTCATGGTGTGCCTGTCCTGGTGTCTTTGTCTTTCTGGGGGCTCGTCCCTTGGGCCACCCCTGCTTCCCCACTAAGAGCTGTACTTCCTTGTGCCTGCAGGAGAAGAAGGATTATCAGCAGAAGCTGGCAAAGGAGCAGGGCTCTCTCAG ......................................................................................................................................................................................................((.(((((.(((......)))..............(((((((((.....)))))).))).........))))).))((.((((......))))..)).................................................... .....................................................................................................................................................................................................198................................................................................................297................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................CGGTCTTGCTTTCTTCTATCCTGGAGAA.................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |