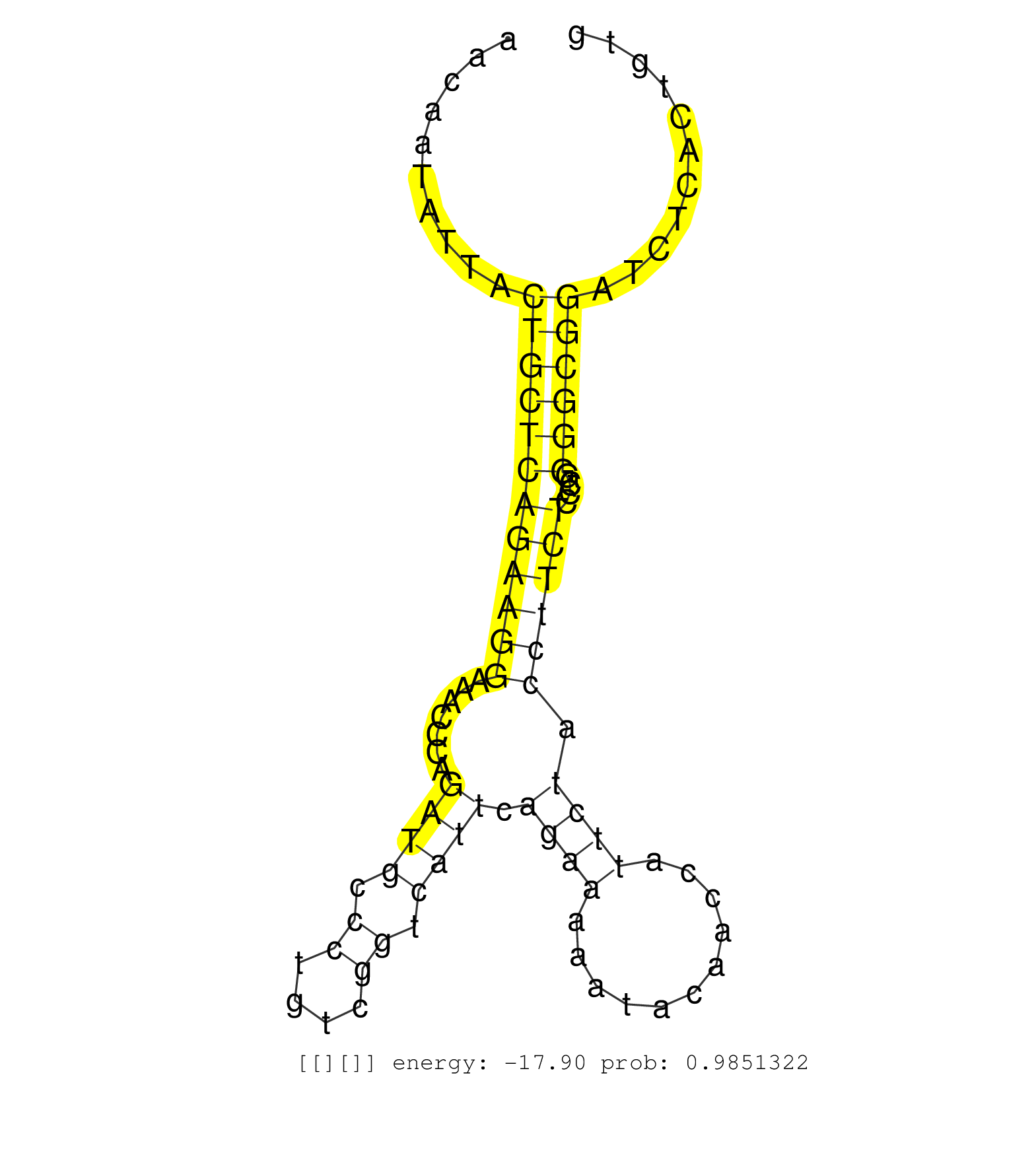

| Gene: A130092J06Rik | ID: uc008jej.1_intron_2_0_chr2_31907736_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CACTGGTGGTGATCCCGCCCAAGCACCGGTAACAGCCAGGCTGCTGCGAGGTGAGACTCAACGTAGTAACAATATTACTGCTCAGAAGGAAACCCAGATGCCCTGTCGGTCATTCAGAAAAATACAACCATTCTACCTTCTCCCGGGGCGGATCTCACTGTGCTAAAACCGGCCGGCCTCCTGGCTCCTCCACGTGCTTCCCTGCAGGGAGGGGGGAGCCAGGGACAACAGGCGCCCACCCACCCCACCC .............................................................................((((((((((((.......((((.((....)).)))).((((...........)))).))))))....))))))................................................................................................... ...................................................................68............................................................................................162...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TATTACTGCTCAGAAGGAAACCCAGAT....................................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - |
| ..........................................................................................................................TACAACCATTCTACCTTCTCCCGGGGC..................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TCGGTCATTCAGAAAAATACAACCATTC..................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TATTACTGCTCAGAAGGAAACCCAGATG...................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................TGCTCAGAAGGAAACCCAGATGCCCT.................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................TAACAGCCAGGCTGCTGCGAGGTGA.................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................TAACAGCCAGGCTGCTGCGAGGgga.................................................................................................................................................................................................... | 25 | gga | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................ACAATATTACTGCTCAGAAGGAAACCCAGA........................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................TCTCCCGGGGCGGATCTCAC............................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TCTCACTGTGCTAAAACCGGCCGGCCT....................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTACTGCTCAGAAGGAAACCCAGATG...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................TACTGCTCAGAAGGAAACCCAGATG...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................TAACAATATTACTGCTCAGAAGGAAACCC........................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................ACAATATTACTGCTCAGAAGGAAACCC........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TCAGAAGGAAACCCAGATGCCCTGTCGG............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................TGCGAGGTGAGACTCAACGTAGTAACA................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................TAGTAACAATATTACTGCTCAGAAGGA................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................TGCTCAGAAGGAAACCCAGATGCCCTG................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................TATTACTGCTCAGAAGGAAACCCAG......................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TCTCCCGGGGCGGATCTCACTGTGC....................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TAGTAACAATATTACTGCTCAGAAGG................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................CTCACTGTGCTAAAACCGGCCGGCCTCCT.................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGTCGGTCATTCAGAAAAATACAACCAT....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................TATTACTGCTCAGAAGGAAACCCAGATGC..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................TACCTTCTCCCGGGGCGGATCTCACT........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACTGGTGGTGATCCCGCCCAAGCACCGGTAACAGCCAGGCTGCTGCGAGGTGAGACTCAACGTAGTAACAATATTACTGCTCAGAAGGAAACCCAGATGCCCTGTCGGTCATTCAGAAAAATACAACCATTCTACCTTCTCCCGGGGCGGATCTCACTGTGCTAAAACCGGCCGGCCTCCTGGCTCCTCCACGTGCTTCCCTGCAGGGAGGGGGGAGCCAGGGACAACAGGCGCCCACCCACCCCACCC .............................................................................((((((((((((.......((((.((....)).)))).((((...........)))).))))))....))))))................................................................................................... ...................................................................68............................................................................................162...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|