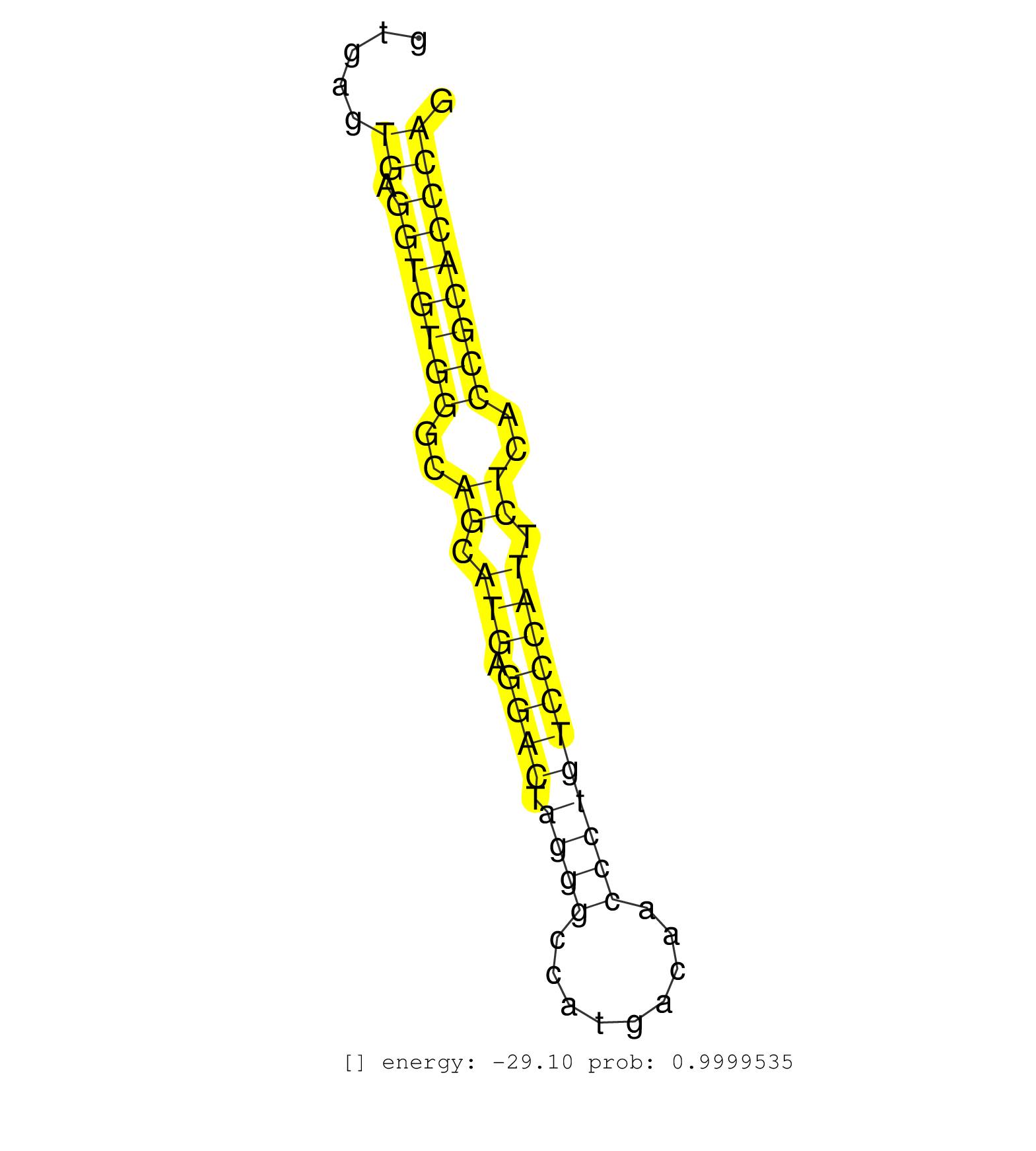
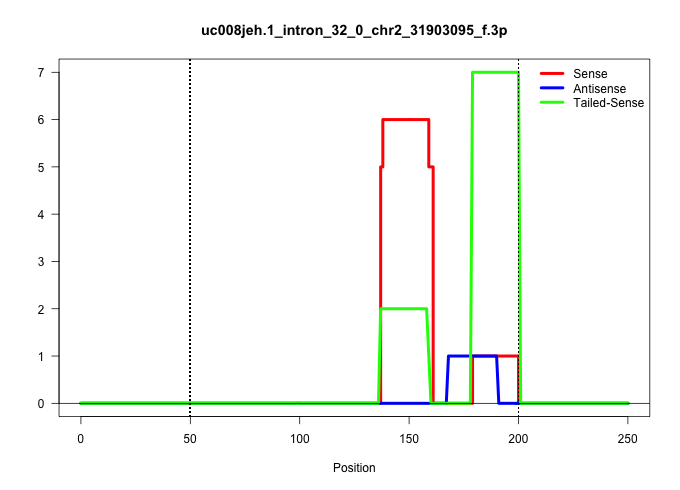
| Gene: Nup214 | ID: uc008jeh.1_intron_32_0_chr2_31903095_f.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) PIWI.ip |
(18) TESTES |
| TTTCTGATGTCTTCCTTCTCAGAGCCACGGTAAAGGCTTACTCTGGGCTAGAAGCCGGTTTCCTCTGAGTCTGTTGACTCGGTCCAAGCTTGTCACCACAGCTCCTGTGTGGTACATCCAAGACAGGTGACAGTGAGTGAGGTGTGGGCAGCATGAGGACTAGGGCCATGACAACCCTGTCCCATTCTCACCGCACCCAGGTTCGGGAGCAGCGGCAATACTGCATCCTTCGGTACCCTCGCCAGTCAGA .........................................................................................................................................((.(((((((..((.(((.((((.((((.........))))))))))).))..)))))))))................................................... ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................TGAGGTGTGGGCAGCATGAGGt........................................................................................... | 22 | t | 8.00 | 3.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCATTCTCACCGCACCCAGt................................................. | 22 | t | 7.00 | 1.00 | - | - | - | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - |
| .........................................................................................................................................TGAGGTGTGGGCAGCATGAGGACT......................................................................................... | 24 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - |
| .........................................................................................................................................TGAGGTGTGGGCAGCATGAGGAC.......................................................................................... | 23 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGAGGTGTGGGCAGCATGAGG............................................................................................ | 21 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TGAGGTGTGGGCAGCATGAGGA........................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................TGTGGGCAGCATGAGGACTAGGG..................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCATTCTCACCGCACCCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................TGAGGTGTGGGCAGCATGAGGAa.......................................................................................... | 23 | a | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GAGGTGTGGGCAGCATGAGGACT......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCCCATTCTCACCGCACCCAGa................................................. | 22 | a | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTTCTGATGTCTTCCTTCTCAGAGCCACGGTAAAGGCTTACTCTGGGCTAGAAGCCGGTTTCCTCTGAGTCTGTTGACTCGGTCCAAGCTTGTCACCACAGCTCCTGTGTGGTACATCCAAGACAGGTGACAGTGAGTGAGGTGTGGGCAGCATGAGGACTAGGGCCATGACAACCCTGTCCCATTCTCACCGCACCCAGGTTCGGGAGCAGCGGCAATACTGCATCCTTCGGTACCCTCGCCAGTCAGA .........................................................................................................................................((.(((((((..((.(((.((((.((((.........))))))))))).))..)))))))))................................................... ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................TGACAACCCTGTCCCATTCTCAC........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................CTCGCCAGTCAGAct | 15 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................GTCACCACAGCTCCT................................................................................................................................................ | 15 | 15 | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |