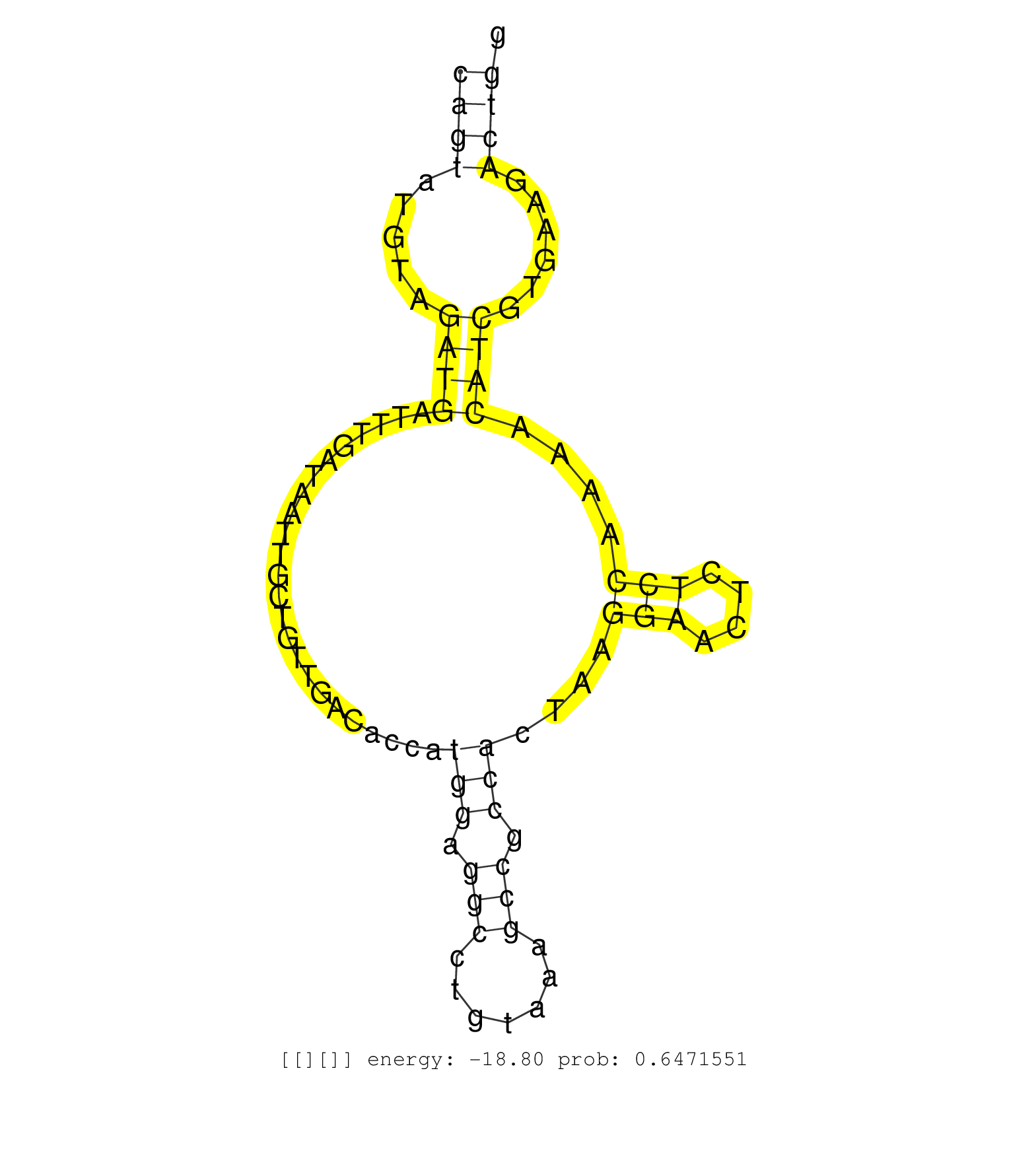

| Gene: 2610205E22Rik | ID: uc008jcu.1_intron_1_0_chr2_30669276_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(12) TESTES |
| GTTCAGAAACTCCCCTAGCATCTTCCGTGAGATTCTCCATGAGGATTTGGGTGAGTACAGCAAGCCCACCCAAATGCCAGTCCGATGCAGTATGTAGATGATTTGATAATTGCTGTTGACACCATGGAGGCCTGTAAAGCCGCCACTAAGGAACTCTCCAAAACATCGTGAAGACTGGAGTACCATGTGTCTGCCAAGAAAGTTCAGCTCTGCCAACTCTAGTAACCTACCTAGAGTACATACTTGAGGG .......................................................................................((((.....((((........................(((.(((.......))).)))....(((....)))....))))......))))......................................................................... .......................................................................................88........................................................................................178...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TAAGGAACTCTCCAAAACATCGTGAAGA............................................................................ | 28 | 1 | 4.00 | 4.00 | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................TGTAGATGATTTGATAATTGCTGTTGAC.................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................TAAGGAACTCTCCAAAACATCGTGAAG............................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| .......................................................................................................TGATAATTGCTGTTGACACCATGGAGGCC...................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TAAGGAACTCTCCAAAACATCGTGAAGAC........................................................................... | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TACCATGTGTCTGCCAAGAAAGTTCAGC.......................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................TAAGGAACTCTCCAAAACATCGTGAAGACT.......................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TCTGCCAAGAAAGTTCAGCTCTGCCAACTC............................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................TGATAATTGCTGTTGACACCATGGAGGC....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................ATAATTGCTGTTGACACCATGGAGGCC...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TGTGTCTGCCAAGAAAGTTCAGCcc........................................ | 25 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................TGCCAGTCCGATGCAGTATGTAGATGATT................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TAGTAACCTACCTAGAGTACATACTTG.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................TAATTGCTGTTGACACCATGGAGGCC...................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGCCAGTCCGATGCAGTATGTAGA........................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................TGATTTGATAATTGCTGTTGACACCATG............................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................TGCCAGTCCGATGCAGTATGTAGATG...................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................TGTAGATGATTTGATAATTGCTGTTGACA................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GTTCAGAAACTCCCCTAGCATCTTCCGTGAGATTCTCCATGAGGATTTGGGTGAGTACAGCAAGCCCACCCAAATGCCAGTCCGATGCAGTATGTAGATGATTTGATAATTGCTGTTGACACCATGGAGGCCTGTAAAGCCGCCACTAAGGAACTCTCCAAAACATCGTGAAGACTGGAGTACCATGTGTCTGCCAAGAAAGTTCAGCTCTGCCAACTCTAGTAACCTACCTAGAGTACATACTTGAGGG .......................................................................................((((.....((((........................(((.(((.......))).)))....(((....)))....))))......))))......................................................................... .......................................................................................88........................................................................................178...................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................CAAAACATCGTGAAaaac.............................................................................. | 18 | aaac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |