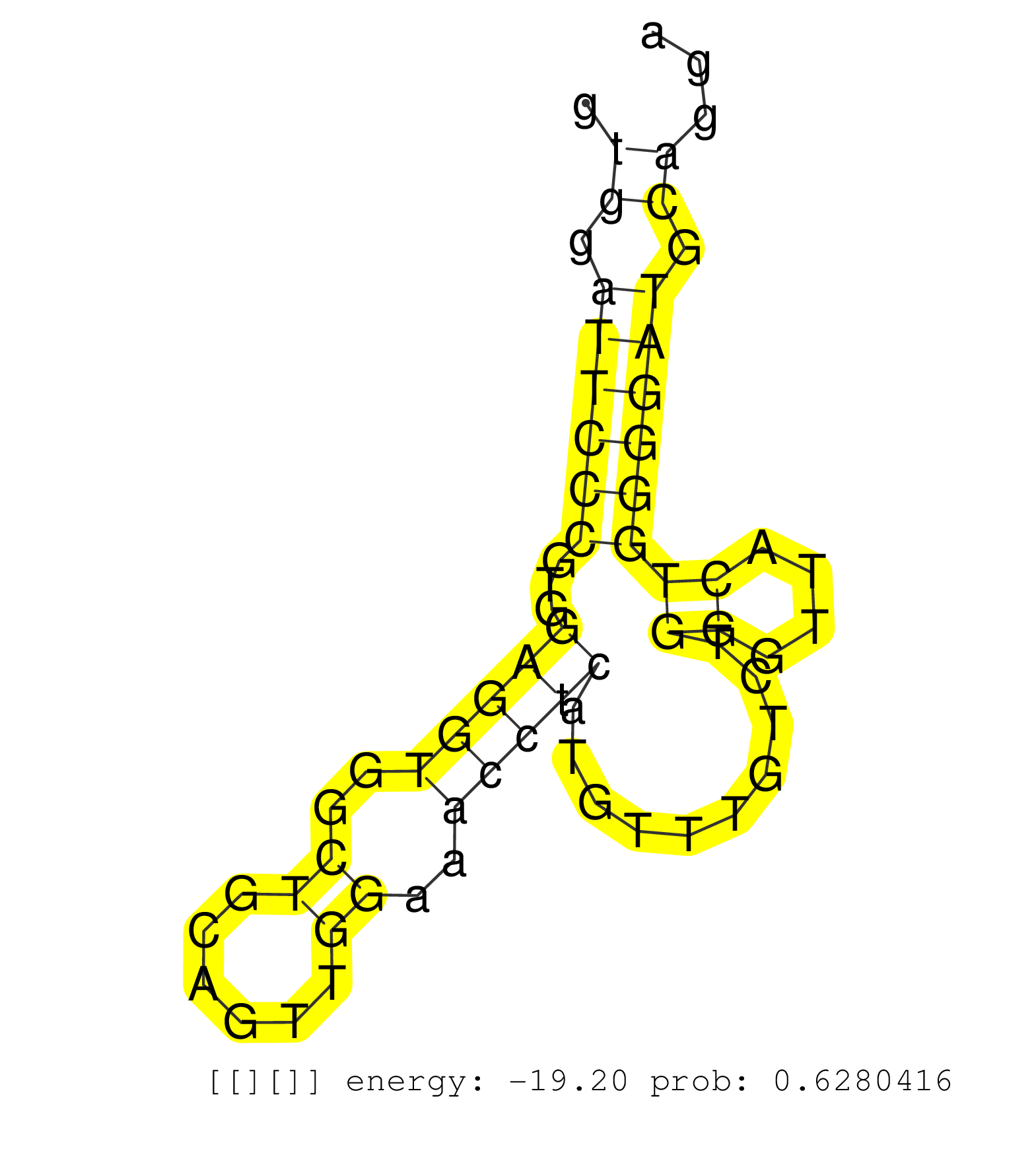

| Gene: Ppp2r4 | ID: uc008jcn.1_intron_8_0_chr2_30298918_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| TGCTGAGTATGGGTGCCTTTGAGTGGGAGGTGGGGATGAGGTGAACGAGATCACCTTCCTCCTAGGCCGGGGGCTGCTCTCCAGCACAGGTGGATTCCCGTGGAGGTGGCTGCAGTTGGAAACCTCATGTTTGTCTGGGTTACTGGGGATGCAGGAGGCGAGACAGGCCTCACCTCCCTGGTTTTCCCCTCCTTTCTCAGTGCCTGGAGAAGTTCCCTGTGATCCAGCACTTCAAGTTCGGGAGCCTGCT ..........................................................................................((.((((((...(((((..((......))..)))))..........((....)))))))).))................................................................................................. .........................................................................................90................................................................156............................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TGTTTGTCTGGGTTACTGGGGATGC.................................................................................................. | 25 | 1 | 24.00 | 24.00 | 9.00 | 10.00 | 2.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGGATTCCCGTGGAGGTGGC............................................................................................................................................ | 20 | 1 | 5.00 | 5.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGTTTGTCTGGGTTACTGGGGATG................................................................................................... | 24 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...TGAGTATGGGTGCCTTTGAGTGGGA.............................................................................................................................................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGTTTGTCTGGGTTACTGGGGATGCA................................................................................................. | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGTTACTGGGGATGCAGGAGGC........................................................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TCCAGCACTTCAAGTTCGGGAGCCTG.. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - |
| ..................................................................................................................................TTGTCTGGGTTACTGGGGATGCAGGA.............................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAGTATGGGTGCCTTTGAGTGGG............................................................................................................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTCCCGTGGAGGTGGCTGCAGTTGG................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................................................................................TTCAAGTTCGGGAGCCTGCT | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .GCTGAGTATGGGTGCCTTTGA.................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................TTGTCTGGGTTACTGGGGATGCAGca.............................................................................................. | 26 | ca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGCAGTTGGAAACCTCATGTTTGTCTGG................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................TGTTTGTCTGGGTTACTGGGGATGa.................................................................................................. | 25 | a | 1.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGCAGTTGGAAACCTCATGTTTGTCTG................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....GAGTATGGGTGCCTTTGAGTGGGAGG............................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................TCTCAGTGCCTGGAGAAGTTCCCTGT.............................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGTGCCTGGAGAAGTTCCCTGTG............................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGAGTATGGGTGCCTTTGAGTGGGAag............................................................................................................................................................................................................................ | 27 | ag | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TGTCTGGGTTACTGGGGATGCAGGAGG............................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGGATTCCCGTGGAGGTGGCTGCAGT...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...TGAGTATGGGTGCCTTTGAGTGGGg.............................................................................................................................................................................................................................. | 25 | g | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGGCTGCAGTTGGAAACCTC............................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................................TGTGATCCAGCACTTCAAGTTCGGGAGCCTG.. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................TGGGTTACTGGGGATGCAGGAGGCGt......................................................................................... | 26 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TACTGGGGATGCAGGAGGCGAGA....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCTGAGTATGGGTGCCTTTGAGTGGGAGGTGGGGATGAGGTGAACGAGATCACCTTCCTCCTAGGCCGGGGGCTGCTCTCCAGCACAGGTGGATTCCCGTGGAGGTGGCTGCAGTTGGAAACCTCATGTTTGTCTGGGTTACTGGGGATGCAGGAGGCGAGACAGGCCTCACCTCCCTGGTTTTCCCCTCCTTTCTCAGTGCCTGGAGAAGTTCCCTGTGATCCAGCACTTCAAGTTCGGGAGCCTGCT ..........................................................................................((.((((((...(((((..((......))..)))))..........((....)))))))).))................................................................................................. .........................................................................................90................................................................156............................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................TTCCCCTCCTTTCTCAGTGCCTGGA.......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |