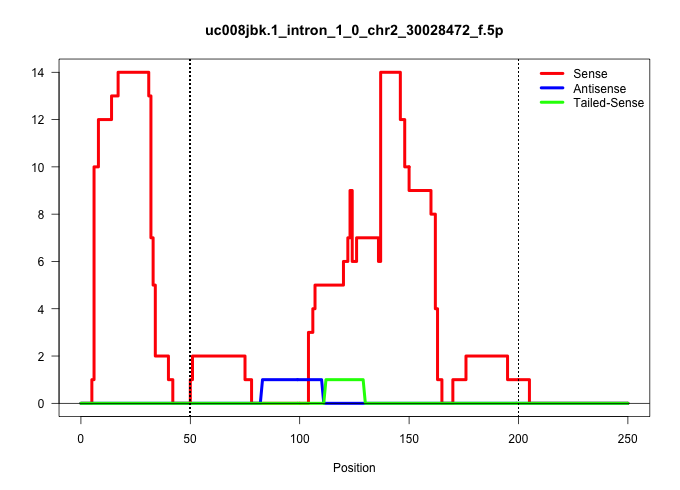
| Gene: Endog | ID: uc008jbk.1_intron_1_0_chr2_30028472_f.5p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(16) TESTES |
| CGAACTTACCAAAATGTCTATGTCTGCACGGGGCCGCTTTTCCTGCCCAGGTAAGCCTGTAGCTGACGGGGCCTCCGACTCCTGTTCTCCAACCTTGGGCTTTCTGTGTCACTGTAGCCAGAGACCTTTGGGTGGCCTAGAACTCTGCTGTGCCCTCAGGTGCCTGGCCCCACACCTGCCTTTGCTCCCCACGTCTGACCTGCCTTACCACCTGTAACCCGTGGCTGCACGGGAGGCTGTCACCCTTGCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......TACCAAAATGTCTATGTCTGCACGGG.......................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | - | 1.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................TAGAACTCTGCTGTGCCCTCAGGTG........................................................................................ | 25 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TGTGTCACTGTAGCCAGAGA.............................................................................................................................. | 20 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TAGAACTCTGCTGTGCCCTCAGGTGC....................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - |
| ...........................................................................................................................ACCTTTGGGTGGCCTAGAACTCT........................................................................................................ | 23 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........CCAAAATGTCTATGTCTGCACGGGGC........................................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......TACCAAAATGTCTATGTCTGCACGGGG......................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GACCTTTGGGTGGCCTAGAACTCTGC...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TTACCAAAATGTCTATGTCTGCACGGG.......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......TACCAAAATGTCTATGTCTGCACGGGGC........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............TGTCTATGTCTGCACGGGGCCGCTTT.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGCCTGTAGCTGACGGGGCCTCCGA............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TGTAGCCAGAGACCggcc........................................................................................................................ | 18 | ggcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................TAGAACTCTGCTGTGCCCTCAGGTGCCT..................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCTAGAACTCTGCTGTGCCCTCAGG.......................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGTCACTGTAGCCAGAGACCTTTGGGTGG................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......TACCAAAATGTCTATGTCTGCACGG........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGCCTGTAGCTGACGGGGCCTC............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGCCTTTGCTCCCCACGTCTGACCTGCCT............................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................CTATGTCTGCACGGGGCCGCTTTTC................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GAGACCTTTGGGTGGCCTAGAACTCTGCTG.................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................CACACCTGCCTTTGCTCCCCACGTC....................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................GTCACTGTAGCCAGAGACCTTTGGGTGGC.................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................TTTGGGTGGCCTAGAACTCTGC...................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| CGAACTTACCAAAATGTCTATGTCTGCACGGGGCCGCTTTTCCTGCCCAGGTAAGCCTGTAGCTGACGGGGCCTCCGACTCCTGTTCTCCAACCTTGGGCTTTCTGTGTCACTGTAGCCAGAGACCTTTGGGTGGCCTAGAACTCTGCTGTGCCCTCAGGTGCCTGGCCCCACACCTGCCTTTGCTCCCCACGTCTGACCTGCCTTACCACCTGTAACCCGTGGCTGCACGGGAGGCTGTCACCCTTGCC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................GTTCTCCAACCTTGGGCTTTCTGTGTCA........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |