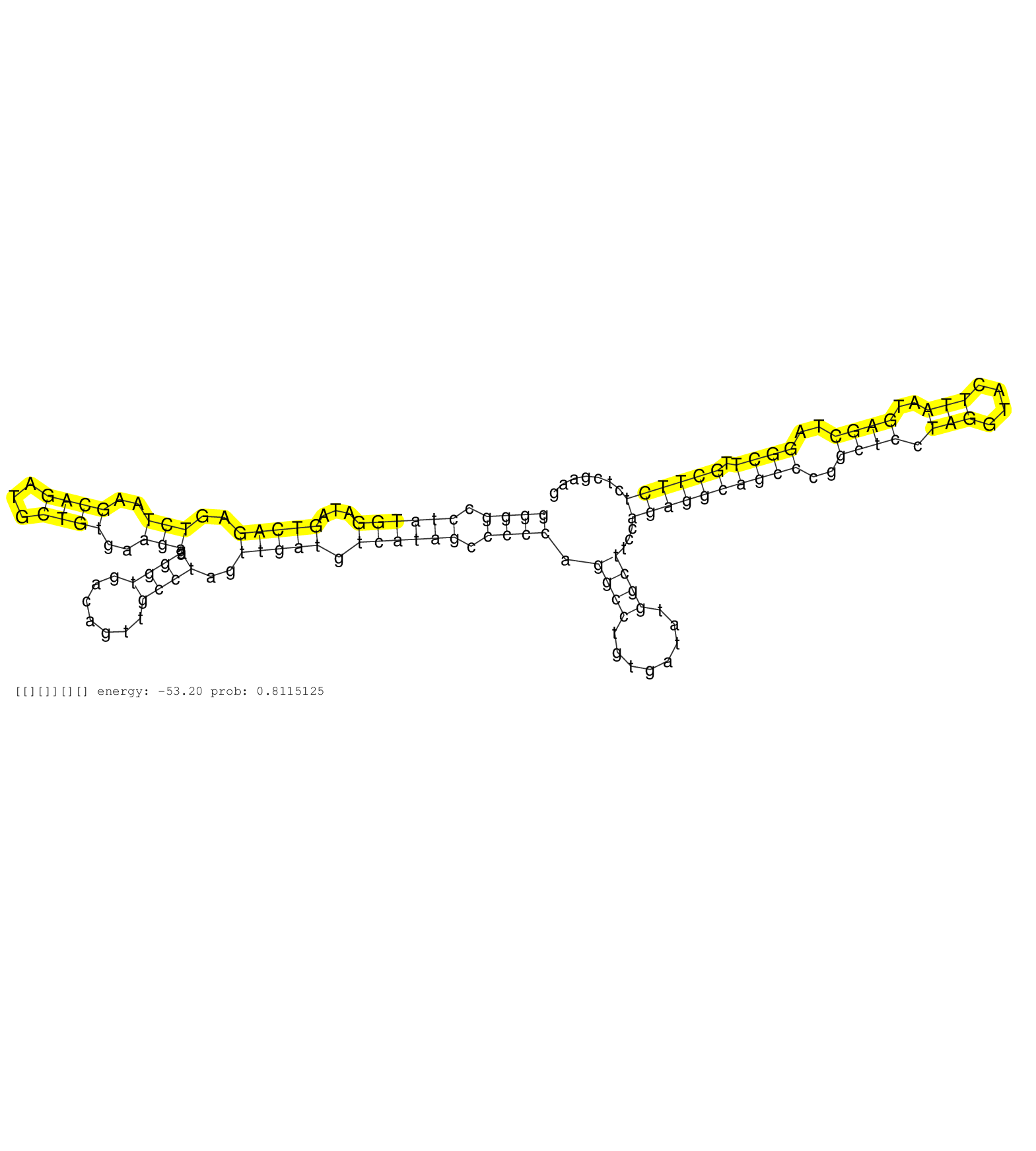

| Gene: Zer1 | ID: uc008jbf.1_intron_13_0_chr2_29966853_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| GGGACTTGCTGGTGATCACCCTGAAGACCACCTTTAGATTCCTATGCCCTGGGGCCTATGGATAGTCAGAGTCTAAGCAGATGCTGTGAAGAGAGGTGACAGTTGCCTAGTTGATGTCATAGCCCCCAGGCCTGTGATATGGCTTCCAGAGGCAGCCCGGCTCCTAGGTACTTAATGAGCTAGGCTTGCTTCTCTCGAAGGTATGTGGAGCTGGTCAGTGCTGCCTGCACTTTTGAGCCACACGAGACTT ..................................................((((.((((((...(((((..(((..((((...))))..))).((((.......))))..))))).)))))).)))).((((........))))...((((((((((..((((.(((....)))..))))..)))).))))))......................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................TGCCTGCACTTTTGAGCCACACGAGACT. | 28 | 1 | 8.00 | 8.00 | 4.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................TGGATAGTCAGAGTCTAAGCAGATGCTG.................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................TGGATAGTCAGAGTCTAAGCAGATGC...................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................TATGGATAGTCAGAGTCTAAGCAGATG....................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................TATGGATAGTCAGAGTCTAAGCAGATGC...................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TAGGTACTTAATGAGCTAGGCTTGCTTC.......................................................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................TGGATAGTCAGAGTCTAAGCAGATGCTGT................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TAGGTACTTAATGAGCTAGGCTTGCTTCT......................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGCCTGCACTTTTGAGCCACACGAGAC.. | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGCCTGCACTTTTGAGCCACACGAGA... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGCCTAGTTGATGTCATAGCCCCCAG......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................TAAGCAGATGCTGTGAAGAGAGGTGA....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TAGGTACTTAATGAGCTAGGCTTGCTTt.......................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................AGGTACTTAATGAGCaca................................................................... | 18 | aca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................................................GCCTGCACTTTTGAGCCACACGAGA... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................TAGTTGATGTCATAGCCCCCAGGCCTG.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GGGACTTGCTGGTGATCACCCTGAAGACCACCTTTAGATTCCTATGCCCTGGGGCCTATGGATAGTCAGAGTCTAAGCAGATGCTGTGAAGAGAGGTGACAGTTGCCTAGTTGATGTCATAGCCCCCAGGCCTGTGATATGGCTTCCAGAGGCAGCCCGGCTCCTAGGTACTTAATGAGCTAGGCTTGCTTCTCTCGAAGGTATGTGGAGCTGGTCAGTGCTGCCTGCACTTTTGAGCCACACGAGACTT ..................................................((((.((((((...(((((..(((..((((...))))..))).((((.......))))..))))).)))))).)))).((((........))))...((((((((((..((((.(((....)))..))))..)))).))))))......................................................... ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|