
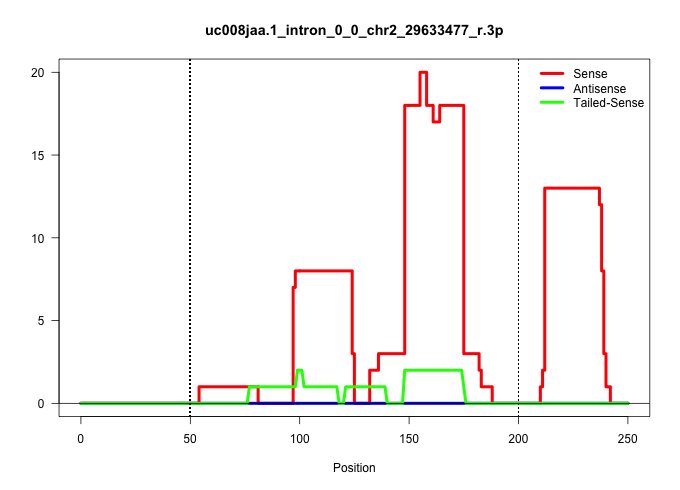
| Gene: Trub2 | ID: uc008jaa.1_intron_0_0_chr2_29633477_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(7) PIWI.ip |
(21) TESTES |
| CAGCCTTTTTACAGCTCAAGCGGGGGGTCCAGAGCTGGGGTCTGCCCCCGCAGGTGCAGGAGGCCTGCAGTCAGCACCACTGCCGTGCTCTCAGCCCTACAGCAAAGCTCAAAGGCATCTCATGAAGGGACATGAGTCCACGTCAGCTTGGTCCCTGCAGCTGTTCCTGACGGTCACGTCTGTGTGTCTGTGACCCACAGAGGTGCAGTGTATGCACGAGACGCAGCAGCAGCTGAGGAAGCTGGTGCAT .............................................................................................(((...(((...((((...(((.(((...(((..((((....))))...)))....))).)))..))))....)))..)))............................................................................ ............................................................................................93....................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TGGTCCCTGCAGCTGTTCCTGACGGTC........................................................................... | 27 | 1 | 15.00 | 15.00 | 1.00 | 5.00 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| .................................................................................................TACAGCAAAGCTCAAAGGCATCTCATG.............................................................................................................................. | 27 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGCACGAGACGCAGCAGCAGCTGAGG............ | 26 | 1 | 4.00 | 4.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................TGCACGAGACGCAGCAGCAGCTGAGGA........... | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TACAGCAAAGCTCAAAGGCATCTCATGA............................................................................................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGAGTCCACGTCAGCTTGGTCCCTGC............................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................TGCACGAGACGCAGCAGCAGCTGAGGAA.......... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................TGCAGGAGGCCTGCAGTCAGCACCACT......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TCCTGACGGTCACGTCTGTGTGTC.............................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TATGCACGAGACGCAGCAGCAGCTGAGGA........... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGCAGCTGTTCCTGACGGTCACGTCTG.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TCCACGTCAGCTTGGTCCCTGCAGC......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TGCACGAGACGCAGCAGCAGCTGAGGAAGC........ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TGCAGCTGTTCCTGACGGTCACGTCTGT................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................TGGTCCCTGCAGCTGTTCCTGACGGgca.......................................................................... | 28 | gca | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ATGAAGGGACATGAGaggt.............................................................................................................. | 19 | aggt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................ATGCACGAGACGCAGCAGCAGCTGAG............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TGGTCCCTGCAGCTGTTCCTGACGGTa........................................................................... | 27 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................GCCCCCGCAGGTGCAGatct........................................................................................................................................................................................... | 20 | atct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CAGCAAAGCTCAAAGGCcc.................................................................................................................................... | 19 | cc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................ACAGCAAAGCTCAAAGGCATCTCATGA............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CACTGCCGTGCTCTCAGCCCTACcg.................................................................................................................................................... | 25 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| CAGCCTTTTTACAGCTCAAGCGGGGGGTCCAGAGCTGGGGTCTGCCCCCGCAGGTGCAGGAGGCCTGCAGTCAGCACCACTGCCGTGCTCTCAGCCCTACAGCAAAGCTCAAAGGCATCTCATGAAGGGACATGAGTCCACGTCAGCTTGGTCCCTGCAGCTGTTCCTGACGGTCACGTCTGTGTGTCTGTGACCCACAGAGGTGCAGTGTATGCACGAGACGCAGCAGCAGCTGAGGAAGCTGGTGCAT .............................................................................................(((...(((...((((...(((.(((...(((..((((....))))...)))....))).)))..))))....)))..)))............................................................................ ............................................................................................93....................................................................................179..................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................CCCTACAGCAAAGCTCAagg........................................................................................................................................... | 20 | agg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................CACTGCCGTGCTCTCaga.............................................................................................................................................................. | 18 | aga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |