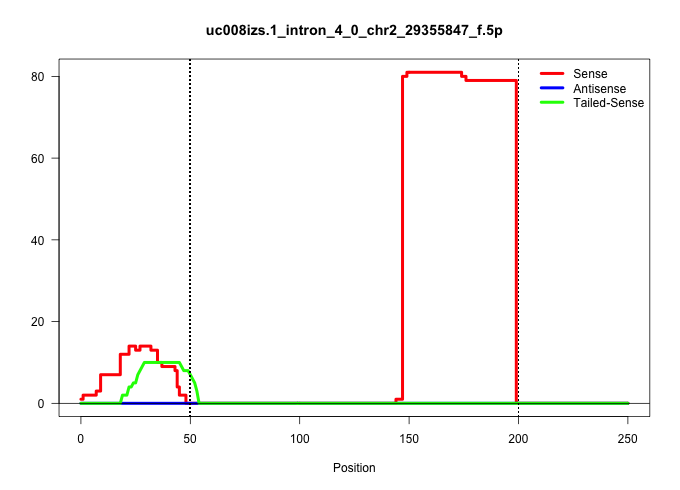
| Gene: Med27 | ID: uc008izs.1_intron_4_0_chr2_29355847_f.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| GCACGATTGTGAAGGGGTATAATGAGAGCGTCTACACAGAGGATGGAAAGGTGAGTGGCCTGGAGTCTTGGGGATAGGTTCTTCCTTACTCTGTCCCAGAACAGAGTTCTAGGTGAGTTGTACTAATAACCAGCAAATCAGCACTGGACTGATCCAGACTAGTCAGCATGAAGAGGGACCCTTGACAAATACTATTTCATTCTCTCCACTTGCCGTTCTAAACCTCTGCTGCTCACAGCAAGCCCGATGC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................ACTGATCCAGACTAGTCAGCATGAAGAGGGACCCTTGACAAATACTATTTCA................................................... | 52 | 1 | 79.00 | 79.00 | 79.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ATAATGAGAGCGTCTACACAGAGGAT.............................................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .........TGAAGGGGTATAATGAGAGCGTCTAC....................................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGAGCGTCTACACAGAGGATGGAA.......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................AGCGTCTACACAGAGGATGGAAAGctg..................................................................................................................................................................................................... | 27 | ctg | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................TGAGAGCGTCTACACAGAGGATGGAAtt........................................................................................................................................................................................................ | 28 | tt | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................GCGTCTACACAGAGGATG............................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .CACGATTGTGAAGGGGTA....................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................TAATGAGAGCGTCTACACAGAGGAT.............................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......TGTGAAGGGGTATAATGAGAGCGTC.......................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................GCGTCTACACAGAGGATGGAAAGctgt.................................................................................................................................................................................................... | 27 | ctgt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................TAATGAGAGCGTCTACACAGAGGATGt............................................................................................................................................................................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................TGATCCAGACTAGTCAGCATGAAGAGG.......................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................ATAATGAGAGCGTCTACACAGAGGATG............................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGGACTGATCCAGACTAGTCAGCATGAAGA............................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........TGAAGGGGTATAATGAGAGCGTCTACAC..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................TAATGAGAGCGTCTACACAGAGGATGtt........................................................................................................................................................................................................... | 28 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................GTCTACACAGAGGATGGAAAGctgg.................................................................................................................................................................................................... | 25 | ctgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGAGCGTCTACACAGAGGATGGAAAGt....................................................................................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................AGAGCGTCTACACAGAGGATGGAAAGct...................................................................................................................................................................................................... | 28 | ct | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................CGTCTACACAGAGGATGGAAAGctgg.................................................................................................................................................................................................... | 26 | ctgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................ATAATGAGAGCGTCTACACAGAGGA............................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GCACGATTGTGAAGGGGTATAATGAGAGCGTCTACACAGAGGATGGAAAGGTGAGTGGCCTGGAGTCTTGGGGATAGGTTCTTCCTTACTCTGTCCCAGAACAGAGTTCTAGGTGAGTTGTACTAATAACCAGCAAATCAGCACTGGACTGATCCAGACTAGTCAGCATGAAGAGGGACCCTTGACAAATACTATTTCATTCTCTCCACTTGCCGTTCTAAACCTCTGCTGCTCACAGCAAGCCCGATGC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................AACCAGCAAATCAGCAtgg........................................................................................................... | 19 | tgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |