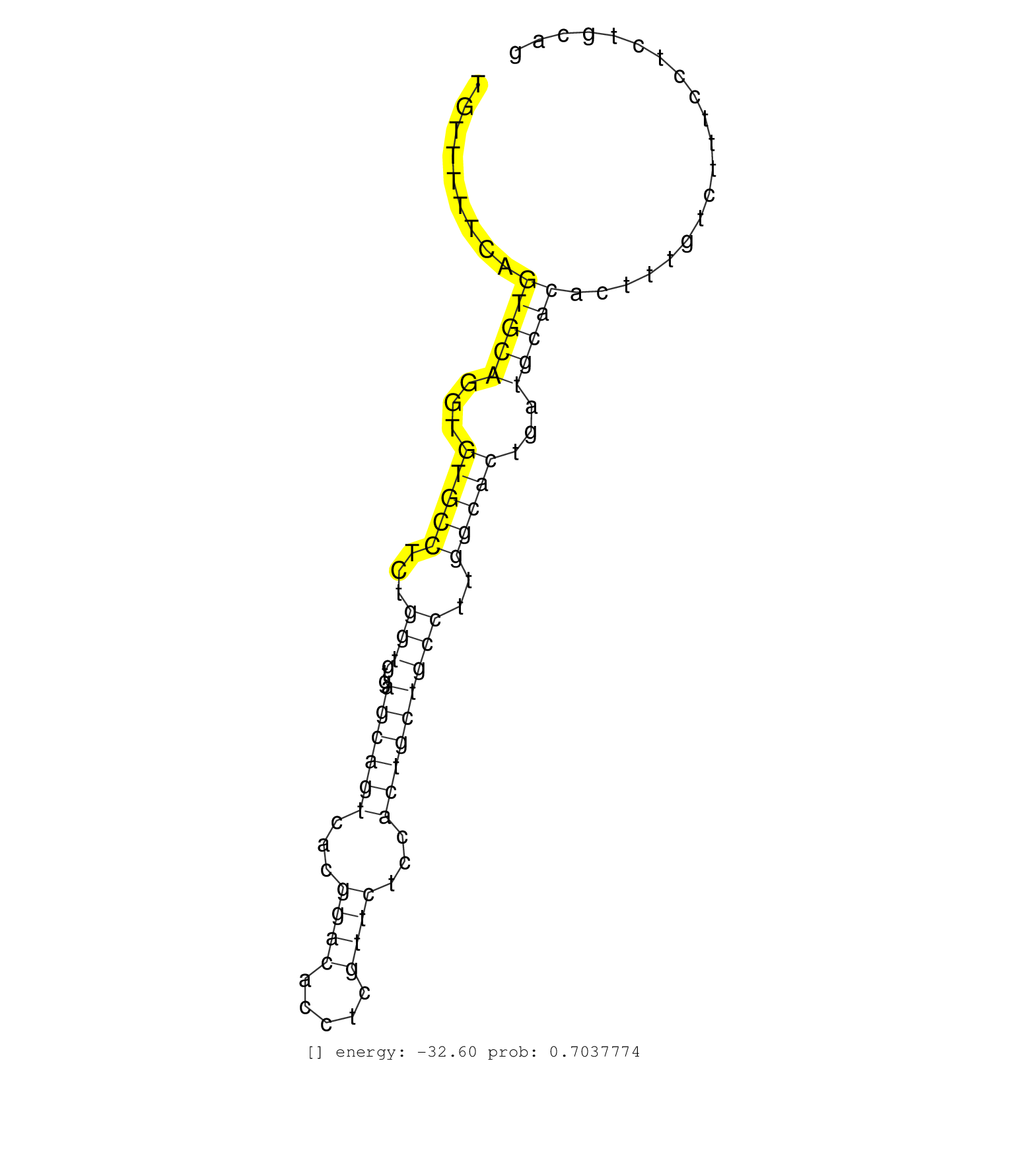

| Gene: Ddx31 | ID: uc008izg.1_intron_1_0_chr2_28700615_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| AAGACTTCATCGCTGTTTAAAAACAACCCTGAGATCCCAGAACTCCACAGGTGTGTTTTTCAGTGCAGGTGTGCCTCTGGTGTGAGCAGTCACGGACACCTCGTTCTCCACTGCTGCCTTGGCACTGATGCACACTTTGTCTTTCCTCTGCAGCACTGTAGTGAAGCAGGCTCGAGAGCAAGTGTTCTCTCCAGAAGCTTTCC ..............................................................(((((...(((((...(((...((((((...((((.....))))...)))))))))..)))))...)))))...................................................................... .....................................................54.................................................................................................153................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGTGTTTTTCAGTGCAGGTGTGCCTC.............................................................................................................................. | 27 | 1 | 18.00 | 18.00 | 6.00 | 5.00 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGTGTTTTTCAGTGCAGGTGTGCCTCT............................................................................................................................. | 28 | 1 | 6.00 | 6.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGTGTTTTTCAGTGCAGGTGTGCCTCT............................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................TAGTGAAGCAGGCTCGAGAGCAAGT.................... | 25 | 2 | 2.00 | 2.00 | - | - | - | - | - | 0.50 | 1.50 | - | - | - | - | - | - |
| ........................................................................................................................................................GCACTGTAGTGAAGCAGGCTCGAGAGC........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................GTAGTGAAGCAGGCTCGAGAGCAAGTGT.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................GTGTGTTTTTCAGTGCAGGTGTGCCT............................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGAAGCAGGCTCGAGAGCAAGTGTT................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......TCATCGCTGTTTAAAAACAACCCTGAGAT........................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGTGTTTTTCAGTGCAGGTGTGCCcc.............................................................................................................................. | 27 | cc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGTGTTTTTCAGTGCAGGTGTGa................................................................................................................................. | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................GCACTGTAGTGAAGCAGGCTCGAGAGCAt...................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................TGAAGCAGGCTCGAGAGCAAGTGTTCTCTC............ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TAGTGAAGCAGGCTCGAGAGCAAG..................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| AAGACTTCATCGCTGTTTAAAAACAACCCTGAGATCCCAGAACTCCACAGGTGTGTTTTTCAGTGCAGGTGTGCCTCTGGTGTGAGCAGTCACGGACACCTCGTTCTCCACTGCTGCCTTGGCACTGATGCACACTTTGTCTTTCCTCTGCAGCACTGTAGTGAAGCAGGCTCGAGAGCAAGTGTTCTCTCCAGAAGCTTTCC ..............................................................(((((...(((((...(((...((((((...((((.....))))...)))))))))..)))))...)))))...................................................................... .....................................................54.................................................................................................153................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) |
|---|