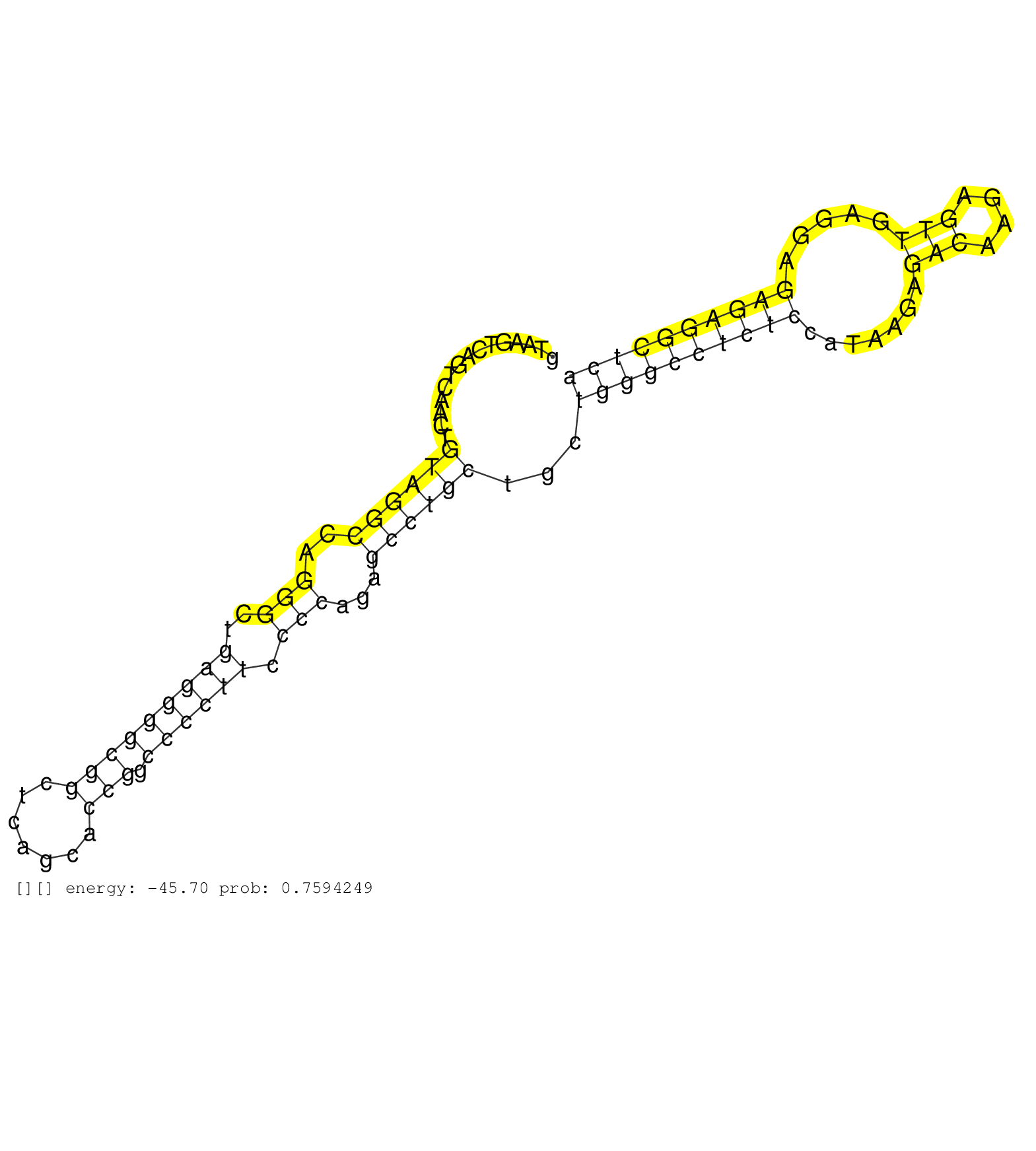

| Gene: Ralgds | ID: uc008iym.1_intron_16_0_chr2_28406084_f.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| GGACGAGCCGGAGGATTATGAGCTGGTGCAGATCATCTCAGAGGATCACAGTAAGTCAGTCAACTGTAGGCCAGGGCTGAGGGGCGGCTCAGCACCGGCCCCTTCCCCAGAGCCTGCTGCTGGGCCTCTCCATAAGAGACAAGAGTTGAGGAGAGAGGCTCATGCGTGGGCCTGTCAGAGAGGACATTGCCTGGTTAAAATAAGAAAGCAGGTACTTCAGGAAGAATGCTCAGTTTAAAATCCACACACG .................................................................((((((..(((..(((((((((.......))).)))))).)))...))))))...((((((((((.......(((....))).....))))))))))........................................................................................ ..................................................51.............................................................................................................162...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................TAAGTCAGTCAACTGTAGGCCAGGGC............................................................................................................................................................................. | 26 | 1 | 9.00 | 9.00 | 2.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - |
| ..................................................GTAAGTCAGTCAACTGTAGGCCAGGGC............................................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | 1.00 | - | - | 2.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ....GAGCCGGAGGATTATGAGCTGGTGCAG........................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................ATAAGAAAGCAGGTgcat................................. | 18 | gcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................AGCTGGTGCAGATCATCTCAGAGGATC........................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................AGTAAGTCAGTCAACTGTAGGCCAGGGC............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGTCAGTCAACTGTAGGCCAGGGCTGt.......................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AATAAGAAAGCAGGTgcat................................. | 19 | gcat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................TAAGAGACAAGAGTTGAGGAGAGAGGC........................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................AGCTGGTGCAGATCATCTC................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................CAGTAAGTCAGTCAACTGTAGGCCAGGGC............................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......GCCGGAGGATTATGAGCTGGTGCAGATC........................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................CAACTGTAGGCCAGGGCTGAGGGGCG.................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGTCAGAGAGGACATTGCCT.......................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............GGATTATGAGCTGGTGCAGATCATCT.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........GGAGGATTATGAGCTGGTGC............................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TAAGAGACAAGAGTTGAGGAGAGAG............................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGCAGATCATCTCAGgga.............................................................................................................................................................................................................. | 18 | gga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................GTGCAGATCATCTCAGAGGATCA.......................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................TGCAGATCATCTCAGAGGATCACAGgtg.................................................................................................................................................................................................... | 28 | gtg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................AAATAAGAAAGCAGGTACTTCAGGAA........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................ATGAGCTGGTGCAGATCA....................................................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CATCTCAGAGGATCA.......................................................................................................................................................................................................... | 15 | 12 | 0.08 | 0.08 | - | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGACGAGCCGGAGGATTATGAGCTGGTGCAGATCATCTCAGAGGATCACAGTAAGTCAGTCAACTGTAGGCCAGGGCTGAGGGGCGGCTCAGCACCGGCCCCTTCCCCAGAGCCTGCTGCTGGGCCTCTCCATAAGAGACAAGAGTTGAGGAGAGAGGCTCATGCGTGGGCCTGTCAGAGAGGACATTGCCTGGTTAAAATAAGAAAGCAGGTACTTCAGGAAGAATGCTCAGTTTAAAATCCACACACG .................................................................((((((..(((..(((((((((.......))).)))))).)))...))))))...((((((((((.......(((....))).....))))))))))........................................................................................ ..................................................51.............................................................................................................162...................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|