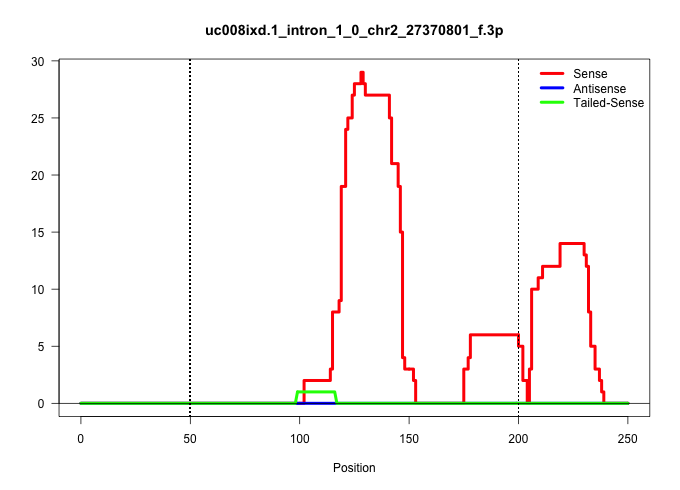
| Gene: Wdr5 | ID: uc008ixd.1_intron_1_0_chr2_27370801_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| TGCATGTAAGTCTGTGTGCCATGTGCATGCAGTCCCTACAGAGGCCAGAAGAGGATGCCAGATCCTCTGAGCTGGACTCACAGATAGTTGTAAGCTCACAAGTGGGAATCAAACTTAGGTCTTCTGGAACTGCATTTGGCGAGCTTAGCTACTGAACTATTTCTCCAACAACCTTTGTTAAAGAGAGGATTTGTTTATAGGCCCCTTACTATAGAGTTCAGCTGACAGCCTCTTCCATTGTGACTCCCCC .....................................................................................................................(((((....((((.....((((.(((..(((........))).))).))))...(((((.......)))))....))))..)))))............................................... ..................................................................................................................115........................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................TCTTCTGGAACTGCATTTGGCGAGCTTA....................................................................................................... | 28 | 1 | 6.00 | 6.00 | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................TTCTGGAACTGCATTTGGCGAGCTTA....................................................................................................... | 26 | 1 | 5.00 | 5.00 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TAGGTCTTCTGGAACTGCATTTGGCGA............................................................................................................ | 27 | 1 | 4.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TCTTCTGGAACTGCATTTGGCGAGCTT........................................................................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGTTAAAGAGAGGATTTGTTTATAGGC................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TACTATAGAGTTCAGCTGACAGCCTCT................. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TACTATAGAGTTCAGCTGACAGCCTC.................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTACTATAGAGTTCAGCTGACAGCCTC.................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................ACTGCATTTGGCGAGCTT........................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................TGGAACTGCATTTGGCGAGCTTAGCTACT................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TACTATAGAGTTCAGCTGACAGCC.................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................GTCTTCTGGAACTGCATTTGGCGAGCT......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGGGAATCAAACTTAGGTCTTCTGGAA......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TAAAGAGAGGATTTGTTTATAGGCCC.............................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................TAAAGAGAGGATTTGTTTATAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GGAACTGCATTTGGCGAGCTTAGCTACT................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TCTTCTGGAACTGCATTTGGCGAGCT......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AAGTGGGAATCAAACgca..................................................................................................................................... | 18 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................TACTATAGAGTTCAGCTGACAGCCTCTTC............... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TATAGAGTTCAGCTGACAGCCTCTTC............... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TACTATAGAGTTCAGCTGACAGCCT................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTAAAGAGAGGATTTGTTTATAGGCCC.............................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGCTGACAGCCTCTTCCAT............ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TAGGTCTTCTGGAACTGCATTTGGCG............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTACTATAGAGTTCAGCTGACAGCCTCT................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................TTAGGTCTTCTGGAACTGCATTTGGCG............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGCTGACAGCCTCTTCCA............. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TCTGGAACTGCATTTGGCGAGCTTAG...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TAGAGTTCAGCTGACAGCCTCTTCCATT........... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGGAACTGCATTTGGCGAGCTTAGCTAC.................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGGGAATCAAACTTAGGTCTTCTGGAAC........................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCATGTAAGTCTGTGTGCCATGTGCATGCAGTCCCTACAGAGGCCAGAAGAGGATGCCAGATCCTCTGAGCTGGACTCACAGATAGTTGTAAGCTCACAAGTGGGAATCAAACTTAGGTCTTCTGGAACTGCATTTGGCGAGCTTAGCTACTGAACTATTTCTCCAACAACCTTTGTTAAAGAGAGGATTTGTTTATAGGCCCCTTACTATAGAGTTCAGCTGACAGCCTCTTCCATTGTGACTCCCCC .....................................................................................................................(((((....((((.....((((.(((..(((........))).))).))))...(((((.......)))))....))))..)))))............................................... ..................................................................................................................115........................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................GTCCCTACAGAGGCgcca............................................................................................................................................................................................................. | 18 | gcca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................TATTTCTCCAACAACtctg.............................................................................. | 19 | tctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |