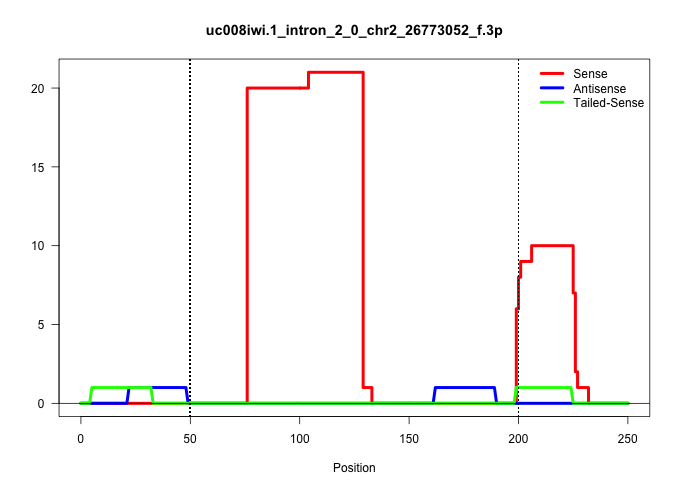
| Gene: Surf2 | ID: uc008iwi.1_intron_2_0_chr2_26773052_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
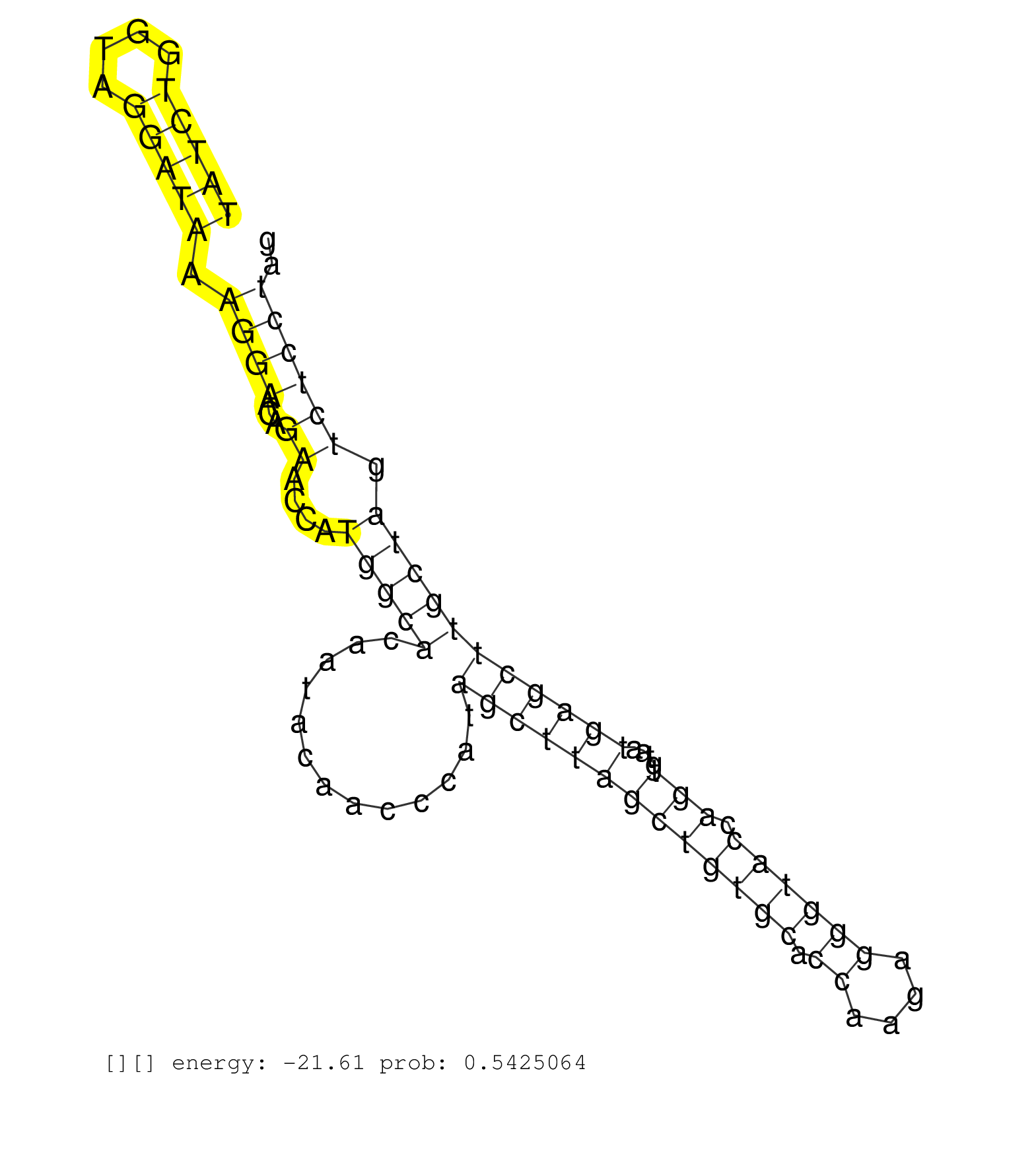
| GACCCTAGTAAGTGCCTTCCCTGAAGCCTCATTCCCGCGCCAAGTTGAAGTGAAGCCAGGCAGTTTCCCCTGAGGTCCACAGCCTTTGGCTGCCCACCCCTATCTGGTAGGATAAAGGAACAGAACCATGGCACAATACAACCCATAGCTTAGCTGTGCACCAAGAGGGTACCAGTGTAATGAGCTTGCTAGTCTCCTAGATGAAGAGTGTCAGAAACAAGGTGTGGAATATGTCCCTGCCTGCCTTCTA ....................................................................................................(((((....))))).((((...((....(((((.............(((((((((((((.((....)))))).)))....))))))))))).)))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................CCACAGCCTTTGGCTGCCCACCCCTATCTGGTAGGATAAAGGAACAGAACCA.......................................................................................................................... | 52 | 1 | 20.00 | 20.00 | 20.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATGAAGAGTGTCAGAAACAAGGTGTG........................ | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATGAAGAGTGTCAGAAACAAGGTGT......................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................AGTGTCAGAAACAAGGTGTGGAATAT.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................ATGAAGAGTGTCAGAAACAAGGTGTG........................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................ATGAAGAGTGTCAGAAACAAGGTGTGG....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................GATGAAGAGTGTCAGAAACAAGGTGc......................... | 26 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................TGGTAGGATAAAGGAACAGAACCATGGCA..................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAAGAGTGTCAGAAACAAGGTGTG........................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....TAGTAAGTGCCTTCCCTGAAGCCTaatt......................................................................................................................................................................................................................... | 28 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| GACCCTAGTAAGTGCCTTCCCTGAAGCCTCATTCCCGCGCCAAGTTGAAGTGAAGCCAGGCAGTTTCCCCTGAGGTCCACAGCCTTTGGCTGCCCACCCCTATCTGGTAGGATAAAGGAACAGAACCATGGCACAATACAACCCATAGCTTAGCTGTGCACCAAGAGGGTACCAGTGTAATGAGCTTGCTAGTCTCCTAGATGAAGAGTGTCAGAAACAAGGTGTGGAATATGTCCCTGCCTGCCTTCTA ....................................................................................................(((((....))))).((((...((....(((((.............(((((((((((((.((....)))))).)))....))))))))))).)))))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................TCCCCTGAGGTCCACAGCCTTTGGCT............................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................GGCACAATACAACCCATAGCTTAttg.................................................................................................. | 26 | ttg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................AAGAGGGTACCAGTGTAATGAGCTTGCT............................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................GAAGCCTCATTCCCGCGCCAAGTTGAA......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |