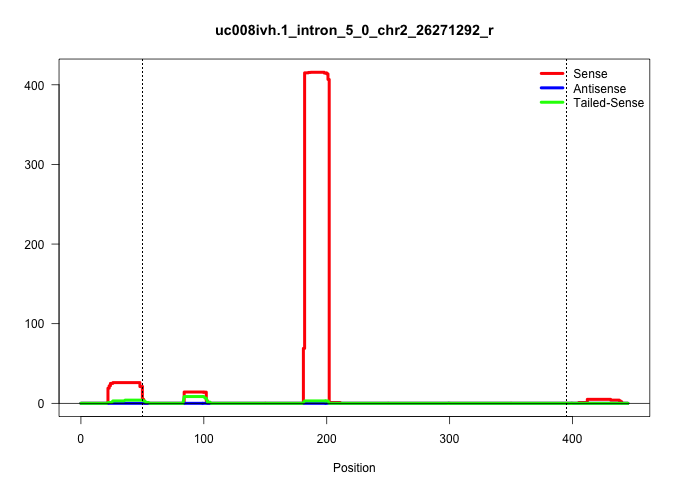
| Gene: Sec16a | ID: uc008ivh.1_intron_5_0_chr2_26271292_r | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(25) TESTES |
| GCCTGCAGGACCACCCACGGCCTCCGTGAATGTGTTTTCTAGAAAAGCAGGTAATAGCTACAGCAGAAATCAGAGGTGTCTGTGTAGAACACTGTCCTTTGGCATCCAGAGCTCAGTCAGCTGTCATGATGCTCACGTTGTCTGTGCATTTTCCACCCTTGAGAGAAAAGCCCAGTTACCCAGTACATGATGAGATCTGAATAGATGGCCCATAGATCTGAGTTTTCTGTTAGTATATATGCAGCCCGTGTGACTGGGTAGAATGGAAACCAAGCCCAATAGTCAATTCTGCTGTGCACCCTCATTAGTTCTTCTGATTCAGTGGGAACCAAATTTGTTGGATGTTGGAGGTCGAATAGCGCTCAACTGAATTTGACTCTCCCTCTCATTGATAGGTGGGTCCAGAGCTCGCTATGTGGATGTTCTAAACCCAAGTGGAACACAG ...............................................................................................................................................................................................................................................................................................................(((((.....)))))(((((((((.((((.....)))).....(((((((((((..((......)).))))))).)))).)))))))))..................................................... .......................................................................................................................................................................................................................................................................................................296................................................................................................395................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................GTACATGATGAGATCTGAAT................................................................................................................................................................................................................................................... | 20 | 1 | 352.00 | 352.00 | 146.00 | 78.00 | 50.00 | 32.00 | 28.00 | - | - | 6.00 | 2.00 | 3.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - |
| .....................................................................................................................................................................................AGTACATGATGAGATCTGAAT................................................................................................................................................................................................................................................... | 21 | 1 | 67.00 | 67.00 | 15.00 | 14.00 | 12.00 | 12.00 | 10.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................TCCGTGAATGTGTTTTCTAGAAAAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 12.00 | 12.00 | - | - | - | - | - | 5.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ......................TCCGTGAATGTGTTTTCTAGAAAAGC............................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 6.00 | 6.00 | - | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................................................................................................AGTACATGATGAGATCTGAA.................................................................................................................................................................................................................................................... | 20 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GTACATGATGAGATCTGAA.................................................................................................................................................................................................................................................... | 19 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CCGTGAATGTGTTTTCTAGAAAAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGAATGTGTTTTCTAGAAAAGCAGGTg........................................................................................................................................................................................................................................................................................................................................................................................................ | 27 | g | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................TCCGTGAATGTGTTTTCTAGAAAAGCAGG.......................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................TATGTGGATGTTCTAAACCCAAGTGGAA..... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TAGAACACTGTCCTTTGG....................................................................................................................................................................................................................................................................................................................................................... | 18 | 2 | 1.50 | 1.50 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................TATGTGGATGTTCTAAACCCAAGTGG....... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GTACATGATGAGATCTGAct................................................................................................................................................................................................................................................... | 20 | ct | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................TATGTGGATGTTCTAAAa............... | 18 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................AGCTCGCTATGTGGATGTTCTAAACC.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................TTCTAGAAAAGCAGGTgggt..................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................AGTACATGATGAGATCTGcat................................................................................................................................................................................................................................................... | 21 | cat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................CGTGAATGTGTTTTCTAGAAAAGCAG........................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GTACATGATGAGATCT....................................................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................TATGTGGATGTTCTAAACCCAAGTGGA...... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TCCGTGAATGTGTTTTCTAGAAAAGCAGt.......................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | t | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................CGTGAATGTGTTTTCTAGAAAAGCAGG.......................................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GTACATGATGAGATCTGA..................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GTACATGATGAGATCTGAATAGATGGCCC.......................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................GTACATGATGAGATCTGAAc................................................................................................................................................................................................................................................... | 20 | c | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGAATGTGTTTTCTAGAAAAGCAGGT......................................................................................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................CGTGAATGTGTTTTCTAGAAAAGCAGGT......................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CATGATGAGATCTGAAT................................................................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TGATGAGATCTGAAT................................................................................................................................................................................................................................................... | 15 | 14 | 0.21 | 0.21 | - | - | 0.14 | - | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCCTGCAGGACCACCCACGGCCTCCGTGAATGTGTTTTCTAGAAAAGCAGGTAATAGCTACAGCAGAAATCAGAGGTGTCTGTGTAGAACACTGTCCTTTGGCATCCAGAGCTCAGTCAGCTGTCATGATGCTCACGTTGTCTGTGCATTTTCCACCCTTGAGAGAAAAGCCCAGTTACCCAGTACATGATGAGATCTGAATAGATGGCCCATAGATCTGAGTTTTCTGTTAGTATATATGCAGCCCGTGTGACTGGGTAGAATGGAAACCAAGCCCAATAGTCAATTCTGCTGTGCACCCTCATTAGTTCTTCTGATTCAGTGGGAACCAAATTTGTTGGATGTTGGAGGTCGAATAGCGCTCAACTGAATTTGACTCTCCCTCTCATTGATAGGTGGGTCCAGAGCTCGCTATGTGGATGTTCTAAACCCAAGTGGAACACAG ...............................................................................................................................................................................................................................................................................................................(((((.....)))))(((((((((.((((.....)))).....(((((((((((..((......)).))))))).)))).)))))))))..................................................... .......................................................................................................................................................................................................................................................................................................296................................................................................................395................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................................................................................................................................................................................................................GATGTTCTAAACCCAgt............ | 17 | gt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |