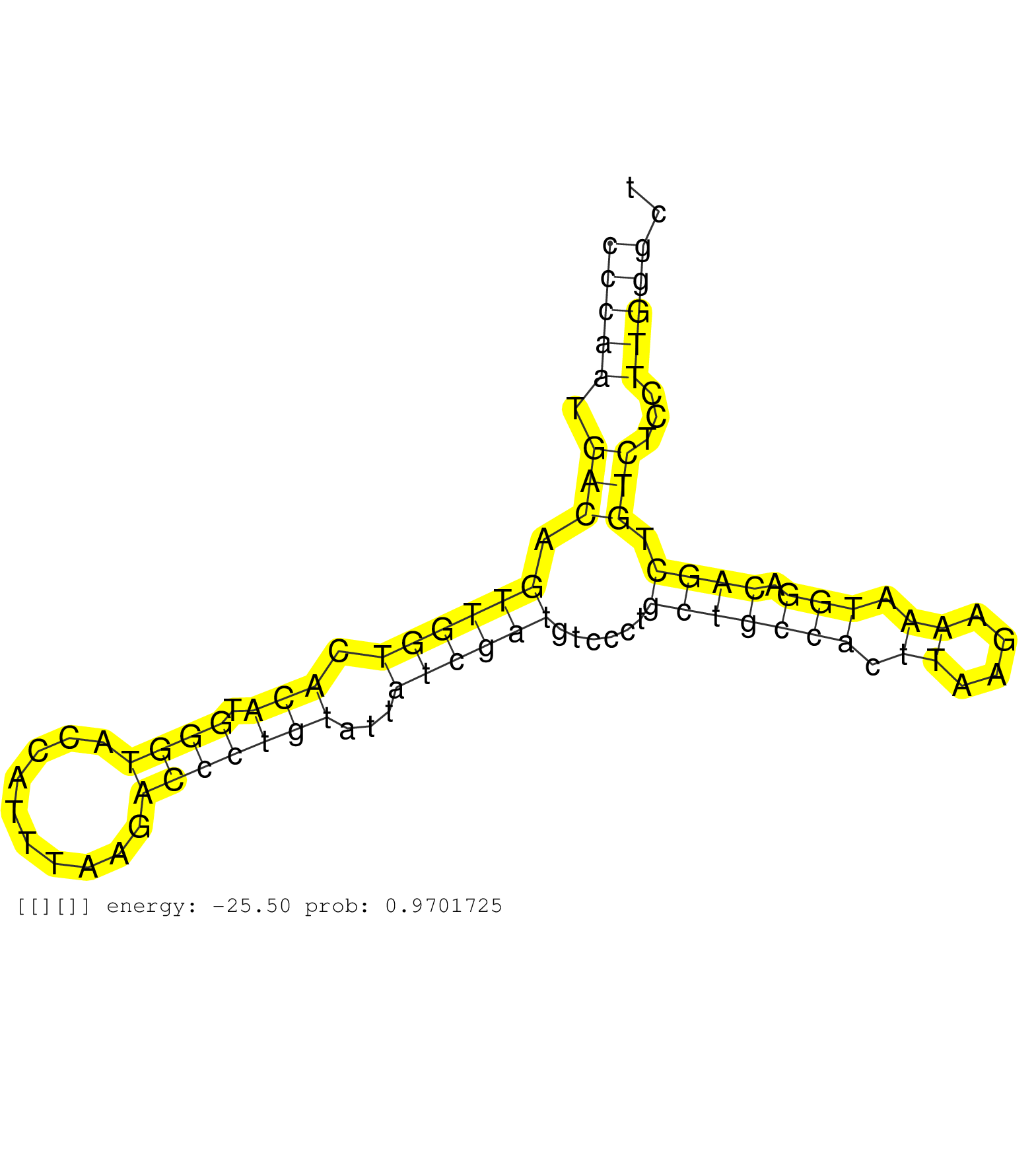
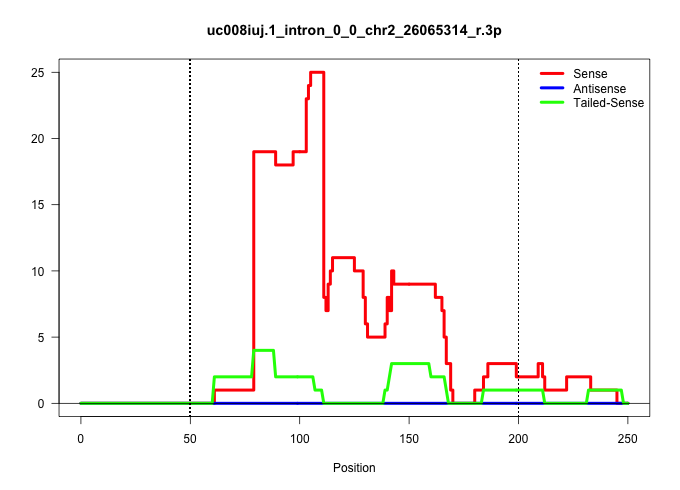
| Gene: Qsox2 | ID: uc008iuj.1_intron_0_0_chr2_26065314_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(1) TDRD1.ip |
(20) TESTES |
| GAGCGCATGTGCGCATGTGTGTGCATGTGTGTACATTCCATTTTGCATATTCTATCTTTTATATTTGCATCCTTCCCAATGACAGTTGGTCACATGGGTACCATTTAAGACCCTGTATTATCGATGTCCCTGCTGCCACTTAAGAAAATGGACAGCTGTCTCCTTGGGCTGTTATTAACCCTCCTTTCATCACATGGCAGGATCAGTTCTGAGAGCCAGACCTTGGCTGGGGCAGATGGCCAGACTCAGC ..........................................................................(((((.(((.((((((.(((.((((..........)))))))...))))))......(((((((.((....)).))).)))).)))...))))).................................................................................. ..........................................................................75.............................................................................................170.............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................TCCTTGGGCTGTTATTAACCCTCCTTTCATCACATGGCAGGATCAGTTCTGA...................................... | 52 | 1 | 67.00 | 67.00 | 67.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TAAGAAAATGGACAGCTGTCTCCTTG.................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................TATTTGCATCCTTCCCAATGACAGTTGt................................................................................................................................................................. | 28 | t | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGTATTATCGATGTCCCTGCTGCCACT.............................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAAATGGACAGCTGTCTCCTTGGGC................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAAGACCCTGTATTATCGATGTCCCTGC..................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AAGAAAATGGACAGCTGga.......................................................................................... | 19 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................TTTAAGACCCTGTATTATCGATGTCCC........................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTTAAGACCCTGTATTATCGATGTCCCT....................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................AGAAAATGGACAGCTGTCTCCTTGGGCT................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGAAAATGGACAGCTGTCTCCTTGGt.................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TAAGAAAATGGACAGCTGTCTCCTT..................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................TGCCACTTAAGAAAATGGACAGCTGTCTC........................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTTAAGACCCTGTATTATCGATGTCCCTGC..................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................GTATTATCGATGTCCCTGCTGCCACTT............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................TGACAGTTGGTCACATGGGTACCATTTt............................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TATTTGCATCCTTCCCAATGACAGTTGG................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TAAGAAAATGGACAGCTGTCTCCTTGG................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTCATCACATGGCAGGATCAGTTCTG....................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TTAAGAAAATGGACAGCTGTCTCCTTGG................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................TTTAAGACCCTGTATTATCGATGTCC......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TATTATCGATGTCCCTGCTGCCACTTAA........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................................TCATCACATGGCAGGATCAGTTCTGA...................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................CAGATGGCCAGACgtc.. | 16 | gtc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TAAGACCCTGTATTATCGATGTCCC........................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTTCATCACATGGCAGGATCAGTTCTGt...................................... | 28 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CTCCTTTCATCACATGGCA................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................GTACCATTTAAGACCCTGTATTATCGAT............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TTAAGACCCTGTATTATCGATGTCC......................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAGAGCCAGACCTTGGCTGGGGC................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTGGCTGGGGCAGATGGCCAGAC..... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................TTAAGAAAATGGACAGCTGTCTCCgtgg................................................................................... | 28 | gtgg | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| GAGCGCATGTGCGCATGTGTGTGCATGTGTGTACATTCCATTTTGCATATTCTATCTTTTATATTTGCATCCTTCCCAATGACAGTTGGTCACATGGGTACCATTTAAGACCCTGTATTATCGATGTCCCTGCTGCCACTTAAGAAAATGGACAGCTGTCTCCTTGGGCTGTTATTAACCCTCCTTTCATCACATGGCAGGATCAGTTCTGAGAGCCAGACCTTGGCTGGGGCAGATGGCCAGACTCAGC ..........................................................................(((((.(((.((((((.(((.((((..........)))))))...))))))......(((((((.((....)).))).)))).)))...))))).................................................................................. ..........................................................................75.............................................................................................170.............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................GTATTATCGATGgcgg............................................................................................................................ | 16 | gcgg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |