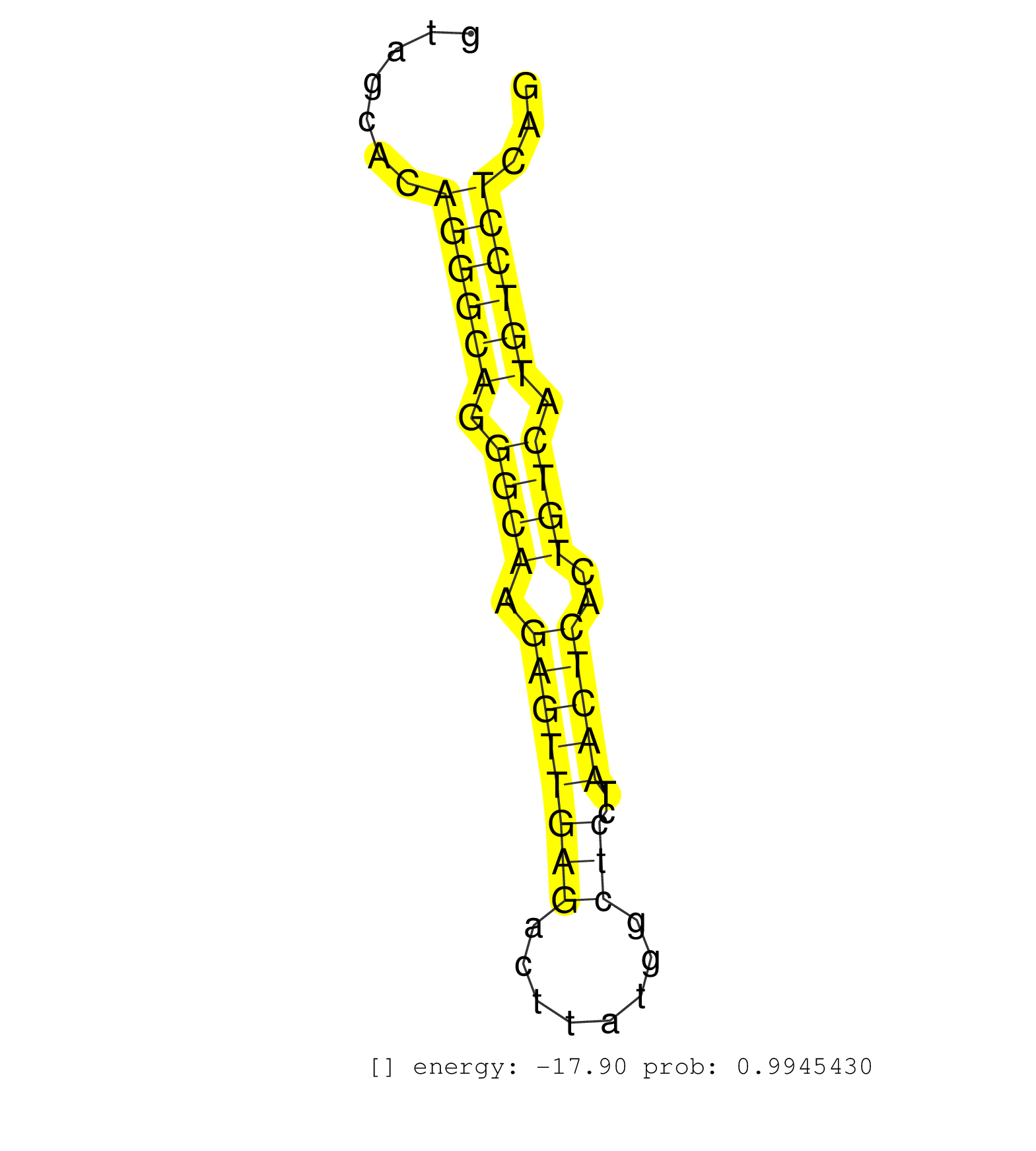

| Gene: Abca2 | ID: uc008isc.1_intron_27_0_chr2_25298273_f | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) PIWI.mut |
(17) TESTES |
| CCCCGTGGCAACTTTATCCCTTATGCCAATGAGGAACGCCAGGAGTACCGGTGAGGCCGTGGGCTGGGGCTGTAGCACAGGGCAGGGCAAGAGTTGAGACTTATGGCTCCTAACTCACTGTCATGTCCTCAGATTACGGCTGTCACCTGATGCCAGCCCCCAGCAGCTGGTGAGCACATTCC ..............................................................................((((((.((((.((((((((........)))..)))))..)))).))))))..................................................... .......................................................................72..........................................................132................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................TAACTCACTGTCATGTCCTCAGt................................................. | 23 | t | 22.00 | 21.00 | - | 14.00 | - | 4.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................TAACTCACTGTCATGTCCTCAG.................................................. | 22 | 1 | 21.00 | 21.00 | 19.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CACAGGGCAGGGCAAGAGTTGAGt................................................................................... | 24 | t | 9.00 | 0.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................CACAGGGCAGGGCAAGAGTTG...................................................................................... | 21 | 1 | 7.00 | 7.00 | 6.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................TAACTCACTGTCATGTCCTCAGA................................................. | 23 | 1 | 6.00 | 6.00 | - | - | 4.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................CAGGGCAGGGCAAGAGTTGAGA................................................................................... | 22 | 1 | 5.00 | 5.00 | - | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................ACAGGGCAGGGCAAGAGTTGAG.................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ............................................................................ACAGGGCAGGGCAAGAGTTGA..................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAACTCACTGTCATGTCCTCAcaa................................................ | 24 | caa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAACTCACTGTCATGTCCTCcgt................................................. | 23 | cgt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGCCGTGGGCTGGGGCTGTA............................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................GAGGAACGCCAGGAGTAC...................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................CCAATGAGGAACGCCAGGAGTACC..................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................TGCCAATGAGGAACGCCAGGAGTACC..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................GGGCAAGAGTTGAGtgag................................................................................ | 18 | tgag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................TAACTCACTGTCATGTCCTCAGAT................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CTAACTCACTGTCATGTCCTCA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................CACAGGGCAGGGCAAGAGTTGt..................................................................................... | 22 | t | 1.00 | 7.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGGGCTGTAGCACAG...................................................................................................... | 15 | 15 | 0.20 | 0.20 | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCCGTGGCAACTTTATCCCTTATGCCAATGAGGAACGCCAGGAGTACCGGTGAGGCCGTGGGCTGGGGCTGTAGCACAGGGCAGGGCAAGAGTTGAGACTTATGGCTCCTAACTCACTGTCATGTCCTCAGATTACGGCTGTCACCTGATGCCAGCCCCCAGCAGCTGGTGAGCACATTCC ..............................................................................((((((.((((.((((((((........)))..)))))..)))).))))))..................................................... .......................................................................72..........................................................132................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|