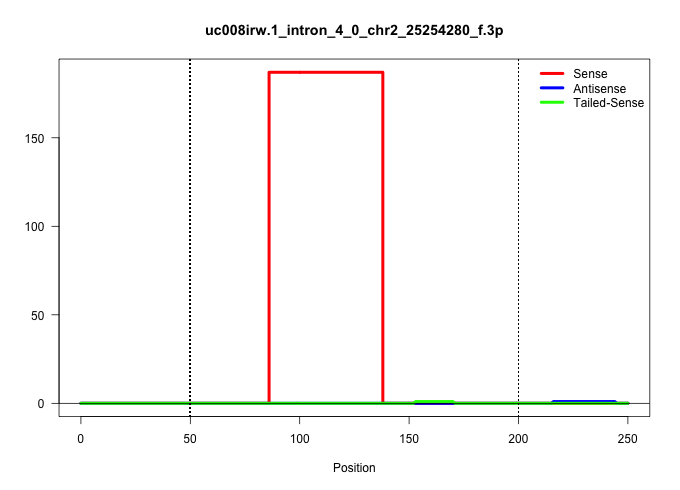
| Gene: Entpd2 | ID: uc008irw.1_intron_4_0_chr2_25254280_f.3p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) TESTES |
| TTATCTCAGAACCTTGAATACCTGGCCAAAAGGTTTGCTGTTTGTCCATCAGGTAATGGTTGCCATGGTTGCTCACAAACACAAAAATGATGTTGCCCAAGTTGGACTTACGAACAGTTCATCAGGCTGCCTCCCTGTGGTGGGGTGGAAGGCAGTGGCTAGAGTGGGGAACTCTATCTGTACTCTCACTAACTCCCCAGATCCATCGCTTCCACCCCTGCTGGCCAAAGGGCTACTCCACCCAAGTGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|
| ......................................................................................ATGATGTTGCCCAAGTTGGACTTACGAACAGTTCATCAGGCTGCCTCCCTGT................................................................................................................ | 52 | 1 | 187.00 | 187.00 | 187.00 | - | - | - |
| .........................................................................................................................................................AGTGGCTAGAGTGGcaga............................................................................... | 18 | caga | 1.00 | 0.00 | - | - | - | 1.00 |
| TTATCTCAGAACCTTGAATACCTGGCCAAAAGGTTTGCTGTTTGTCCATCAGGTAATGGTTGCCATGGTTGCTCACAAACACAAAAATGATGTTGCCCAAGTTGGACTTACGAACAGTTCATCAGGCTGCCTCCCTGTGGTGGGGTGGAAGGCAGTGGCTAGAGTGGGGAACTCTATCTGTACTCTCACTAACTCCCCAGATCCATCGCTTCCACCCCTGCTGGCCAAAGGGCTACTCCACCCAAGTGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................CCTGCTGGCCAAAGGGCTACTCCACCCAA..... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - |
| ..............................................................................................................CAGTTCATCAGGCTGaata......................................................................................................................... | 19 | aata | 1.00 | 0.00 | - | - | 1.00 | - |