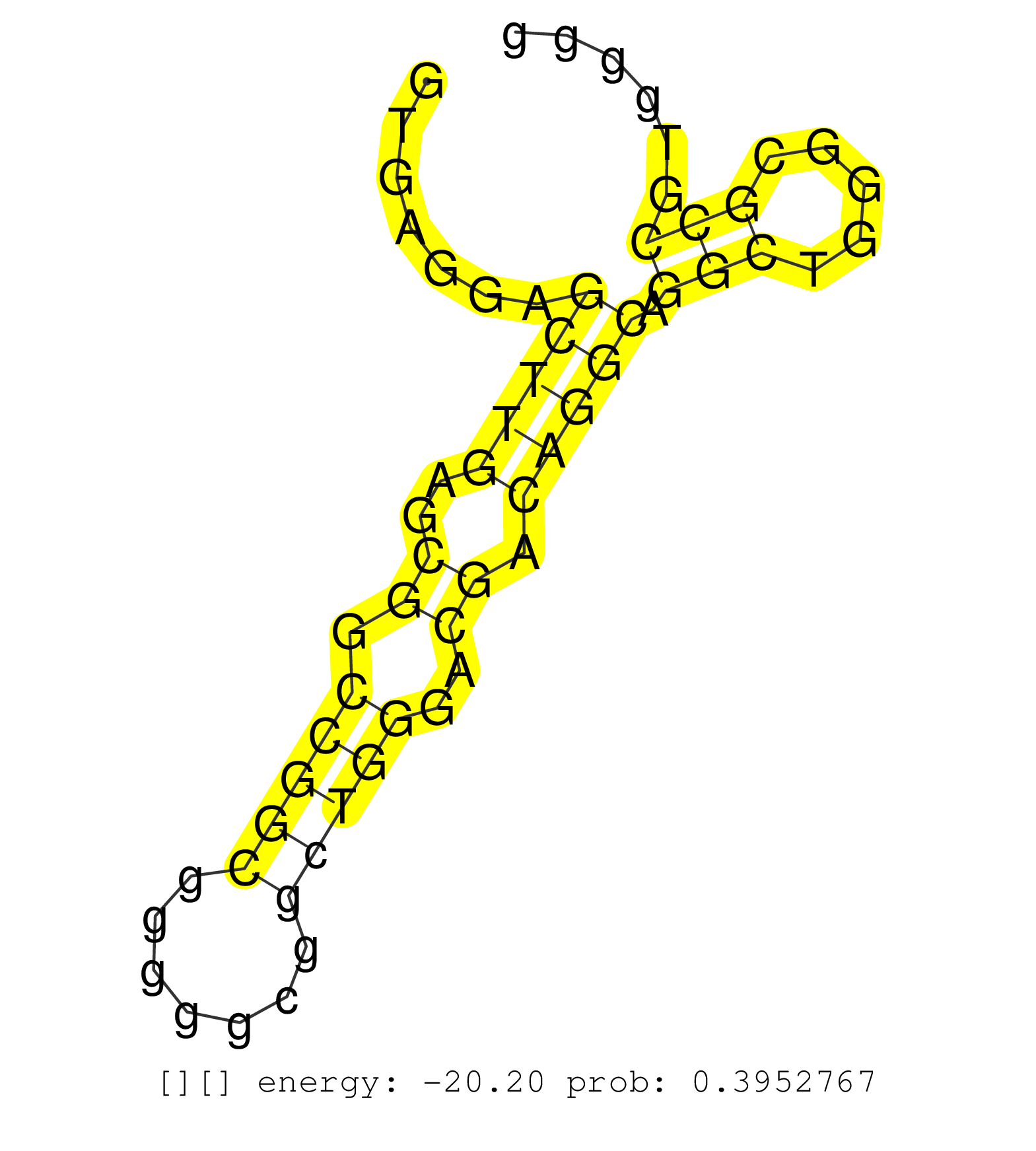

| Gene: Abi1 | ID: uc008ins.1_intron_9_0_chr2_22850211_r.5p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(3) PIWI.mut |
(28) TESTES |
| ACCAGAACCTGACCCGGGTGGCGGACTACTGTGAAAACAACTATATACAGGTGAGGAGCTTGAGCGGCCGGCGGGGGCGGCTGGGACGACAGGCAGGCTGGGCGCCGTGGGGACTGCCCTACTCCGCCACCCTCCGCCCCAGCCCGAGCGGCGGCCGCCGCGGCGATACAGGAAGTGGCCGCCTTGAGAAAATGGGTGAGGAGCAGGGCCGCCGCGGGCGCCGCTGACCCGATGCCGCTCCCCGCAGCCT .........................................................(((((..((.(((((.......)))))..)).))))).(((.....)))................................................................................................................................................ ..................................................51...........................................................112........................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................TGGGACGACAGGCAGGCTGGGCGCCGT.............................................................................................................................................. | 27 | 1 | 24.00 | 24.00 | - | - | - | - | 7.00 | 6.00 | 1.00 | - | 1.00 | - | 4.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................GGGGACTGCCCTACTCCGCCACCCTCCGCCCCAGCCCGAGCGGCGGCCGCCG.......................................................................................... | 52 | 1 | 10.00 | 10.00 | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGAAAACAACTATATACAGGTGAGGAGC............................................................................................................................................................................................... | 28 | 1 | 9.00 | 9.00 | - | - | - | 4.00 | - | - | 2.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TGACCCGATGCCGCTCCCCGCAG... | 23 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGAAAACAACTATATACAGGTGAGGA................................................................................................................................................................................................. | 26 | 1 | 5.00 | 5.00 | - | - | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGAGAAAATGGGTGAGGAGCAGGGCC........................................ | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................TATATACAGGTGAGGAGCTTGAGCGG....................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................CAGGCTGGGCGCCGTGGGGACTGC..................................................................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGGGACGACAGGCAGGCTGGGCGCCGTG............................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........GACCCGGGTGGCGGACT............................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CCAGAACCTGACCCGGGTGGCGGACT............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGAGAAAATGGGTGAGGAGCAGGGC......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................TGAAAACAACTATATACAGGTGAGG.................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................ACAGGTGAGGAGCTTGAGCGGCCGGt.................................................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............GGGTGGCGGACTACTGTGA........................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TGAGAAAATGGGTGAGGAGCAGGGa......................................... | 25 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GGTGGCGGACTACTGTGA........................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGAGCTTGAGCGGCCGG................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TGAAAACAACTATATACAGGTGAGGAG................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TATACAGGTGAGGAGCTTGAGCGGCCGGC.................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGAGCGGCCGGCGGGGGCGGCT........................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TATACAGGTGAGGAGCTT............................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................TGAAAACAACTATATACAGGTGAGGAGCT.............................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............CCGGGTGGCGGACTACTG........................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GACCCGGGTGGCGGACTACTGTGAAA...................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GGGACGACAGGCAGGCTGGGCG.................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGACCCGGGTGGCGGACTACTGTGAA....................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGGGACGACAGGCAGGCTGGGCG.................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................GAAAACAACTATATACAGGTGAGGAGC............................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGGGACGACAGGCAGGCTGGGCGCCG............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGGGACGACAGGCAGG......................................................................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ACCAGAACCTGACCCGGGTGGCGGACTACTGTGAAAACAACTATATACAGGTGAGGAGCTTGAGCGGCCGGCGGGGGCGGCTGGGACGACAGGCAGGCTGGGCGCCGTGGGGACTGCCCTACTCCGCCACCCTCCGCCCCAGCCCGAGCGGCGGCCGCCGCGGCGATACAGGAAGTGGCCGCCTTGAGAAAATGGGTGAGGAGCAGGGCCGCCGCGGGCGCCGCTGACCCGATGCCGCTCCCCGCAGCCT .........................................................(((((..((.(((((.......)))))..)).))))).(((.....)))................................................................................................................................................ ..................................................51...........................................................112........................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................CGCCCCAGCCCGAGCGcg.................................................................................................... | 18 | cg | 7.00 | 0.00 | 4.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CGCCCCAGCCCGAGCGc.................................................................................................... | 17 | c | 6.00 | 0.00 | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CGCCCCAGCCCGAGCG.................................................................................................... | 16 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CGCCCCAGCCCGAGCGcga.................................................................................................... | 19 | cga | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................ACCCTCCGCCCCAGCCCGa........................................................................................................ | 19 | a | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CGCCCCAGCCCGAGCGcgaa.................................................................................................... | 20 | cgaa | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CGCCCCAGCCCGAGCGcgag.................................................................................................... | 20 | cgag | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CGCCCCAGCCCGAGCGGcgaa................................................................................................... | 21 | cgaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTCCGCCCCAGCCCGA....................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CGCCCCAGCCCGAGCGG................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CGCCCCAGCCCGAGCcga..................................................................................................... | 18 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................GCGGACTACTGTacat.......................................................................................................................................................................................................................... | 16 | acat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................GCGGACTACTGTacca.......................................................................................................................................................................................................................... | 16 | acca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............CCCGGGTGGCGGACTACTGTGAAAAC.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................CGCCCCAGCCCGAGCGGCGG................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................CACCCTCCGCCCCAGCCCG........................................................................................................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |