
| Gene: Bmi1 | ID: uc008ilz.1_intron_0_0_chr2_18602103_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.mut |
(11) TESTES |
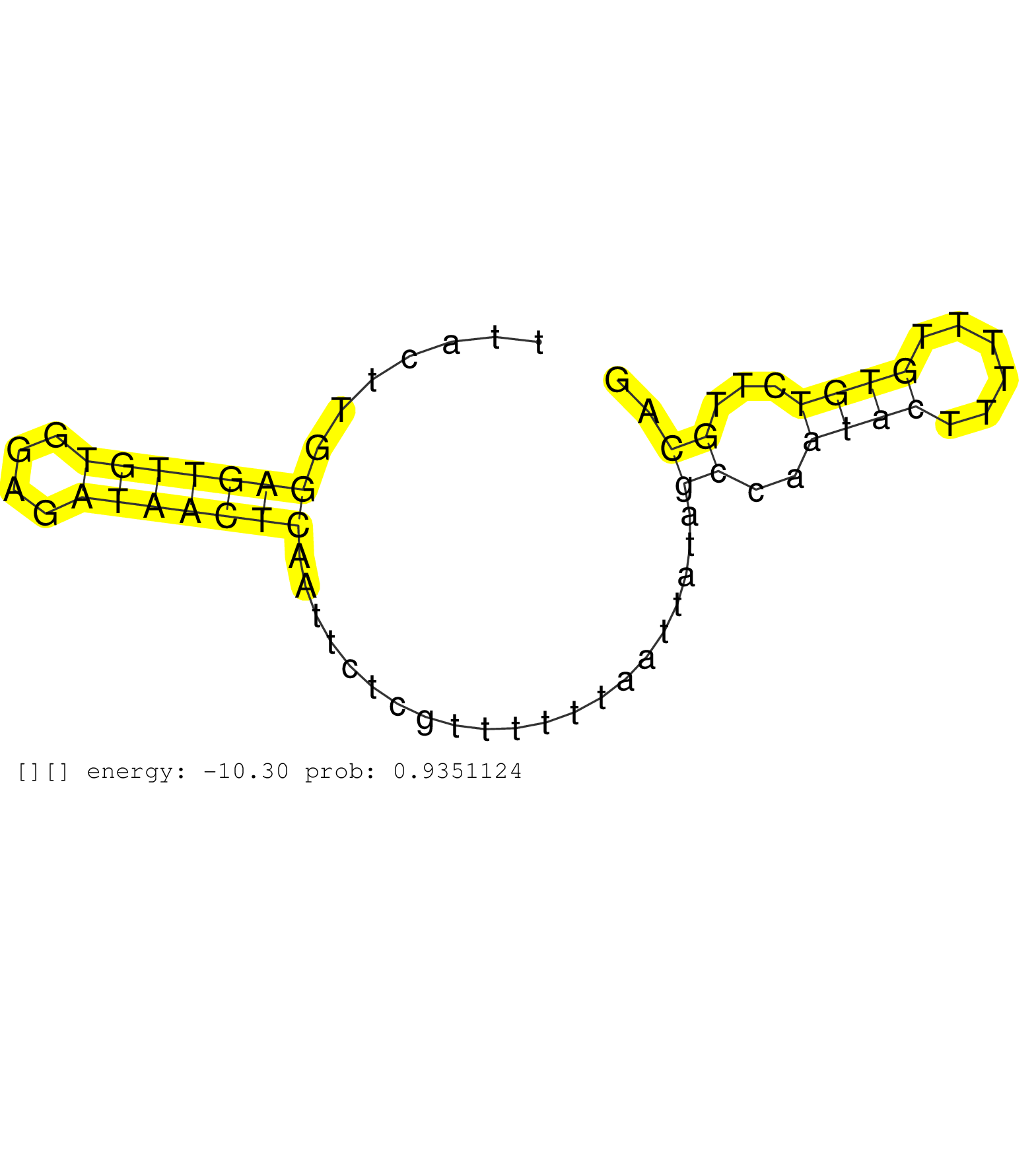
| TTTACACCATTTAACAGTGGAGGTTATTCTGATCTCATTAGTAAATTCCTTTTGTGGAAAACGTTTAGAGAATCCAGCTGTCCAGTGTTAAATGAGTTTTATAAACCGCTCAGCATTCAGGGTATACAGTTACTTGGAGTTGTGGAGATAACTCAATTCTCGTTTTTTAATTATAGCCAATACTTTTTTGTGTCTTGCAGGATCTTTTATCAAGCAGAAATGCATCGAACAACCAGAATCAAGATCACTG ........................................................................................................................................(((((((....))))))).....................((..((((......))))...)).................................................... .................................................................................................................................130...................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TGGAGTTGTGGAGATAACTCAATTCTCGTTTTTTAATTATAGCCAATACTTT................................................................ | 52 | 1 | 22.00 | 22.00 | 22.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GTTGTGGAGATAACTCAA.............................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TTTTTTGTGTCTTGCtct................................................. | 18 | tct | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGATCTTTTATCAAGCAGAAATGC........................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ...................................................................................................................................................................................................................AAGCAGAAATGCATCGAACAACCAGAATC.......... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..........................................................................................................................................................................................................................AATGCATCGAACAACCAGAATCAA........ | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ............................................................................................................................................................................................................................TGCATCGAACAACCAGAATCAAGATCA... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ......................................................................................................................................................................................................................CAGAAATGCATCGAACAACCAGAATCA......... | 27 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .....................................................................................................................................TTGGAGTTGTGGAGA...................................................................................................... | 15 | 11 | 0.18 | 0.18 | - | - | - | - | - | - | - | - | - | - | 0.18 |
| TTTACACCATTTAACAGTGGAGGTTATTCTGATCTCATTAGTAAATTCCTTTTGTGGAAAACGTTTAGAGAATCCAGCTGTCCAGTGTTAAATGAGTTTTATAAACCGCTCAGCATTCAGGGTATACAGTTACTTGGAGTTGTGGAGATAACTCAATTCTCGTTTTTTAATTATAGCCAATACTTTTTTGTGTCTTGCAGGATCTTTTATCAAGCAGAAATGCATCGAACAACCAGAATCAAGATCACTG ........................................................................................................................................(((((((....))))))).....................((..((((......))))...)).................................................... .................................................................................................................................130...................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................AAAACGTTTAGAGAATCCAGCTGTCCA...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GAAATGCATCGAACAAccca.................. | 20 | ccca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |