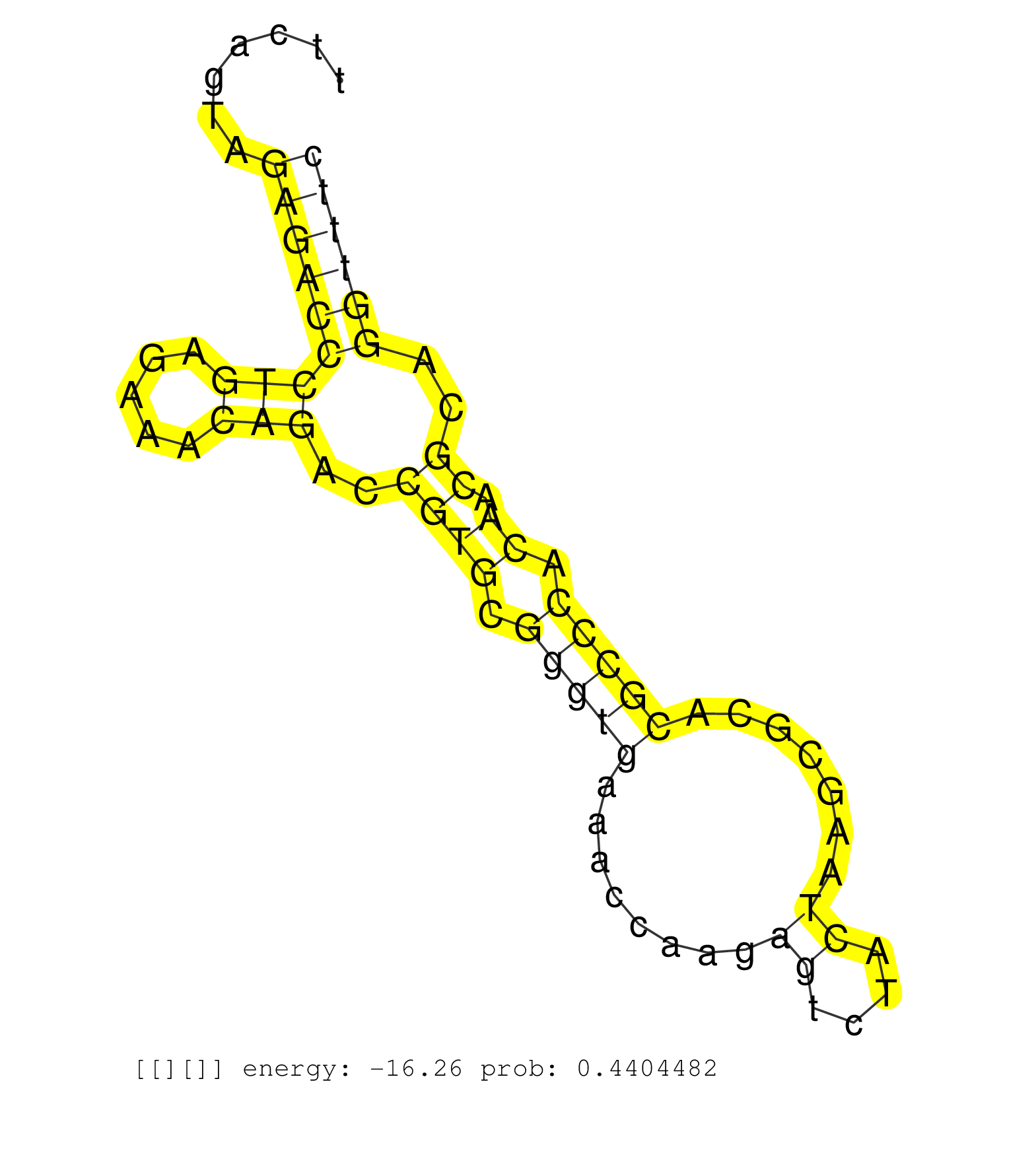
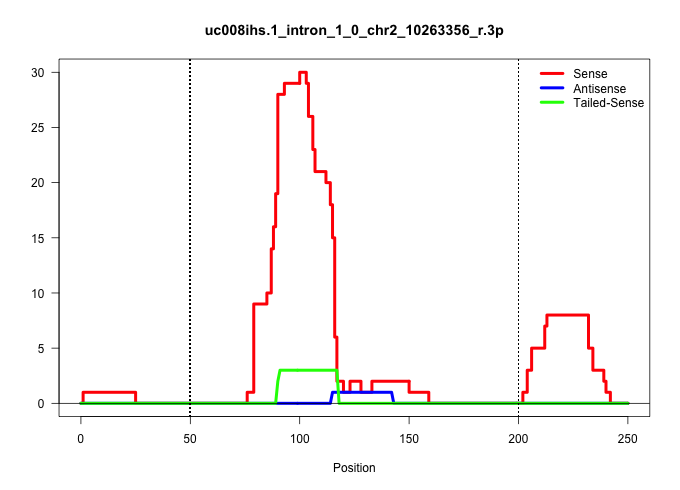
| Gene: AK076687 | ID: uc008ihs.1_intron_1_0_chr2_10263356_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| ATTAAAACTCAGGGCCTTCTGCTTGCACAGCAAACGCTTTCATTACTCAGTCATCTCCCCAGCCCTCGTGCGTCAACACTGAAGCTTCAGTAGAGACCCTGAGAAACAGACCGTGCGGGTGAAACCAAGAGTCTACTAAGCGCACGCCCACAACGCAGGTTTCCTGGGGTAAGTGTCTGATTCTTTATTTTCTTCCGTAGCCTGAGGGAAAATTCAGAAGACAAATACTCCCTTCAACAAACAAGTCAGA ............................................................................................(((((((((.....)))..((((.(((((........((....)).......))))).)).))..))))))....................................................................................... .....................................................................................86...........................................................................163..................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................TAGAGACCCTGAGAAACAGACCGTGC...................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................TAGAGACCCTGAGAAACAGACCGTGCG..................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGAAGCTTCAGTAGAGACCCTGAGA.................................................................................................................................................. | 25 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GTAGAGACCCTGAGAAACAGACCGTGC...................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ...............................................................................TGAAGCTTCAGTAGAGACCCTGAGAAA................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AGGGAAAATTCAGAAGACAAATACTCCC.................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CAGTAGAGACCCTGAGAAACAGACCGTG....................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TAGAGACCCTGAGAAACAGACCGTGCGt.................................................................................................................................... | 28 | t | 2.00 | 4.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGAAGCTTCAGTAGAGACCCTGAGAAAC............................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................GGAAAATTCAGAAGACAAATACTCCCTT................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CAGTAGAGACCCTGAGAAACAGACCGTGC...................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................ACCAAGAGTCTACTAAGCGCACGCCCA.................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................AGAGACCCTGAGAAACAGACCGTGCGt.................................................................................................................................... | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................GAGAAACAGACCGTGCGGGTGAAACCAA.......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................TAGAGACCCTGAGAAACAGACCGTGCGGGT.................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TTCAGTAGAGACCCTGAGAAACAGACC.......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGTAGAGACCCTGAGAAACAGACCGTG....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................TGAGGGAAAATTCAGAAGACAAATACTCCC.................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....................................................................................................................................................................................................................TTCAGAAGACAAATACTCCCTTCAACA........... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................CACTGAAGCTTCAGTAGAGACCCTGAG................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................AGACCCTGAGAAACAGACCGT........................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TTAAAACTCAGGGCCTTCTGCTTG................................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................TTCAGAAGACAAATACTCCCTTCAACAA.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................................TCAGAAGACAAATACTCCCTTCAACAAAC........ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................GTAGAGACCCTGAGAAACAGACCGT........................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTAAGCGCACGCCCACAACGCAGG........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................AGTAGAGACCCTGAGAAACAGACCGTGC...................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATTAAAACTCAGGGCCTTCTGCTTGCACAGCAAACGCTTTCATTACTCAGTCATCTCCCCAGCCCTCGTGCGTCAACACTGAAGCTTCAGTAGAGACCCTGAGAAACAGACCGTGCGGGTGAAACCAAGAGTCTACTAAGCGCACGCCCACAACGCAGGTTTCCTGGGGTAAGTGTCTGATTCTTTATTTTCTTCCGTAGCCTGAGGGAAAATTCAGAAGACAAATACTCCCTTCAACAAACAAGTCAGA ............................................................................................(((((((((.....)))..((((.(((((........((....)).......))))).)).))..))))))....................................................................................... .....................................................................................86...........................................................................163..................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................CGGGTGAAACCAAGAGTCTACTAAGCGC........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGTGTCTGATTCTTTATTTTCTTCCGTA................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |