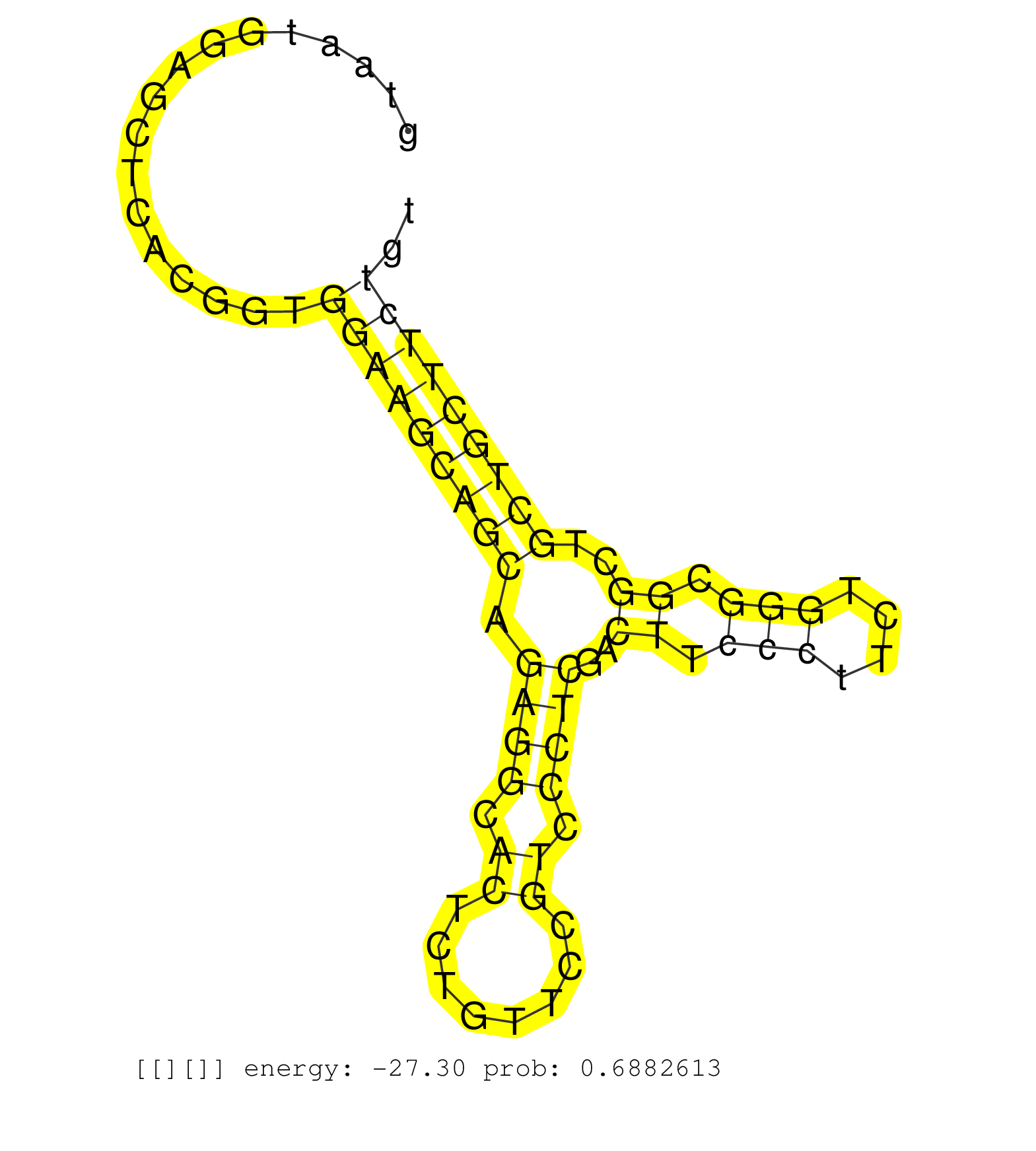
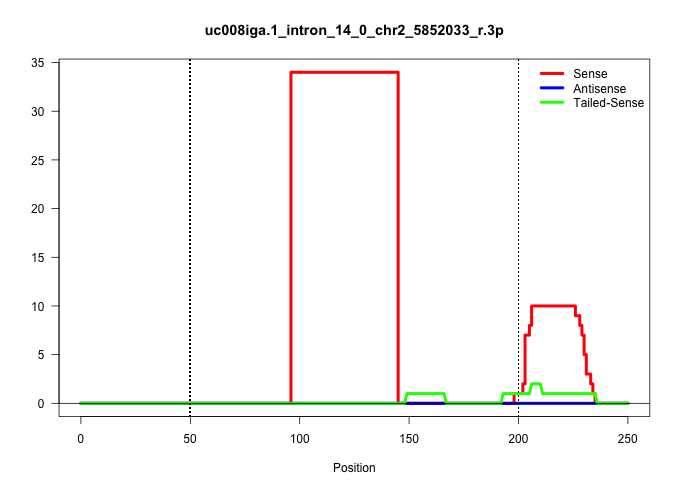
| Gene: Dhtkd1 | ID: uc008iga.1_intron_14_0_chr2_5852033_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(13) TESTES |
| GCTTGTTACGTGGGAGATCCTGGATATAAGTTCCCACAATTGTTTGGTTTTTTTTTAAAGGAACTATTAATAACCAGTGATGATTCTGTTTGTAATGGAGCTCACGGTGGAAGCAGCAGAGGCACTCTGTTCCGTCCCTCGACTTCCCTTCTGGGCGGCTGCTGCTTCTGTGACATGGTCCCTTTGGTTGTCTTTCACAGGACTGTTAAACCTGGGCAAAGAAGCAGCTTCTCTTGAGGAAGTATTGGCC ............................................................................................................(((((((((.((((.((........)).))))..((.(((....))).))..)))))))))................................................................................. ...........................................................................................92.............................................................................171............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................GGAGCTCACGGTGGAAGCAGCAGAGGCACTCTGTTCCGTCCCTCGACTTCCC...................................................................................................... | 52 | 1 | 33.00 | 33.00 | 33.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGACTGTTAAACCTGGGCAAAGAAGC......................... | 28 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGTTAAACCTGGGCAAAGAAGCAGCTT.................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGTTAAACCTGGGCAAAGAAGCAGCTTC................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................TGTAATGGAGCTCACGGTGGAAGCAGC..................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................TCTGGGCGGCTGCTGagc................................................................................... | 18 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................................CTGTTAAACCTGGGCAAAGAAGCAGC...................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TTAAACCTGGGCAAAGAAGCAGCTTCTCT................ | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................TTCACAGGACTGTTAAtt....................................... | 18 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................TGTTAAACCTGGGCAAAGAAGCAGCT..................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TAAACCTGGGCAAAGAAGCAGCTTCTCTTa.............. | 30 | a | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TAAACCTGGGCAAAGAAGCAGCTTCTC................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TAAACCTGGGCAAAGAAGCAGCTTCTCTT............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGTTAAACCTGGGCAAAGAAGCAGC...................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................AGGACTGTTAAACCTGGGCAAAGAAGCA........................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................................GTTAAACCTGGGCAA............................... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GCTTGTTACGTGGGAGATCCTGGATATAAGTTCCCACAATTGTTTGGTTTTTTTTTAAAGGAACTATTAATAACCAGTGATGATTCTGTTTGTAATGGAGCTCACGGTGGAAGCAGCAGAGGCACTCTGTTCCGTCCCTCGACTTCCCTTCTGGGCGGCTGCTGCTTCTGTGACATGGTCCCTTTGGTTGTCTTTCACAGGACTGTTAAACCTGGGCAAAGAAGCAGCTTCTCTTGAGGAAGTATTGGCC ............................................................................................................(((((((((.((((.((........)).))))..((.(((....))).))..)))))))))................................................................................. ...........................................................................................92.............................................................................171............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................CGGTGGAAGCAGCcg..................................................................................................................................... | 15 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |