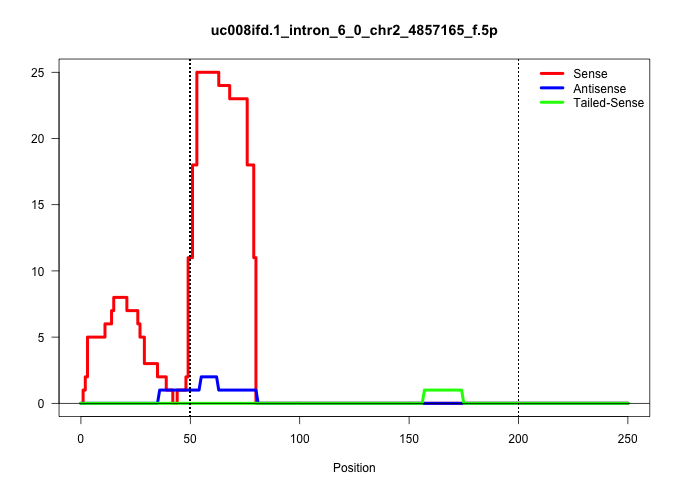
| Gene: Phyh | ID: uc008ifd.1_intron_6_0_chr2_4857165_f.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| CTCTGCTCATCCATGGATCTGGTCGGAACAAAACTCAAGGCTTCCGGAAAGTACGGACTCAGATGGCTACCTCTCCAAAGATGAGTGGTCTAGAACAAAGCCTTCAAGATACATTAAGATAGCAAAGACTGCAGGGCTTCTTACTGAGGGACCAGCTTGTTGACCAAGAAATGCATAAATATTCACTCTGTTAGGAGGAAACAAAATATTATAGGCCCCTAATCACTGAGCCGCGACTGTCCATAGAGTT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................CGGACTCAGATGGCTACCTCTCCAAAG.......................................................................................................................................................................... | 27 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................AGTACGGACTCAGATGGCTACCTCTCCAAA........................................................................................................................................................................... | 30 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TACGGACTCAGATGGCTACCTCTCCAAAG.......................................................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - |
| .................................................AGTACGGACTCAGATGGCTACCTCTCC.............................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...................................................TACGGACTCAGATGGCTACCTCTCC.............................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ...TGCTCATCCATGGATCTGGTCGGAAC............................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ...................................................TACGGACTCAGATGGCT...................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............GATCTGGTCGGAACAAAACTCAAG................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................................TGTTGACCAAGAAATtgt........................................................................... | 18 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................AAGTACGGACTCAGATGGCTACCTCTCC.............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................CGGAAAGTACGGACTCAGA........................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...TGCTCATCCATGGATCTGGTCGGA............................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............GGATCTGGTCGGAACAAAACTCAAGGCT................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..CTGCTCATCCATGGATCTGGTCGG................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........CATGGATCTGGTCGGAACAAAACT....................................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .TCTGCTCATCCATGGATCTG..................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CTCTGCTCATCCATGGATCTGGTCGGAACAAAACTCAAGGCTTCCGGAAAGTACGGACTCAGATGGCTACCTCTCCAAAGATGAGTGGTCTAGAACAAAGCCTTCAAGATACATTAAGATAGCAAAGACTGCAGGGCTTCTTACTGAGGGACCAGCTTGTTGACCAAGAAATGCATAAATATTCACTCTGTTAGGAGGAAACAAAATATTATAGGCCCCTAATCACTGAGCCGCGACTGTCCATAGAGTT |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................GACTCAGATGGCTACCTCTCCAAAGA......................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................AAGGCTTCCGGAAAGTACGGACTCAGA........................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |