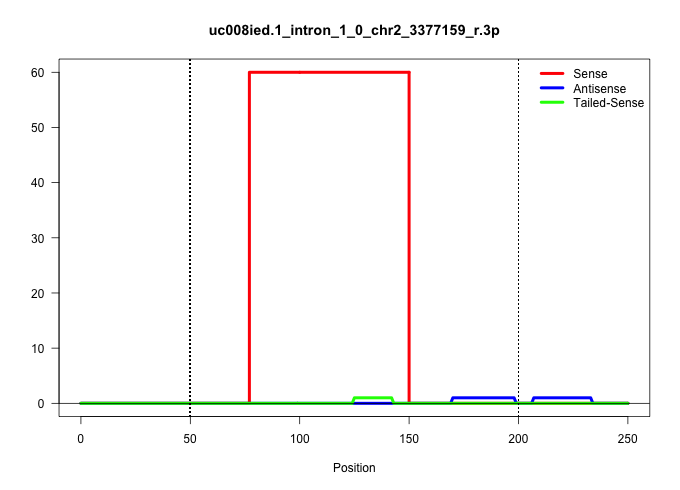
| Gene: Suv39h2 | ID: uc008ied.1_intron_1_0_chr2_3377159_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(7) TESTES |
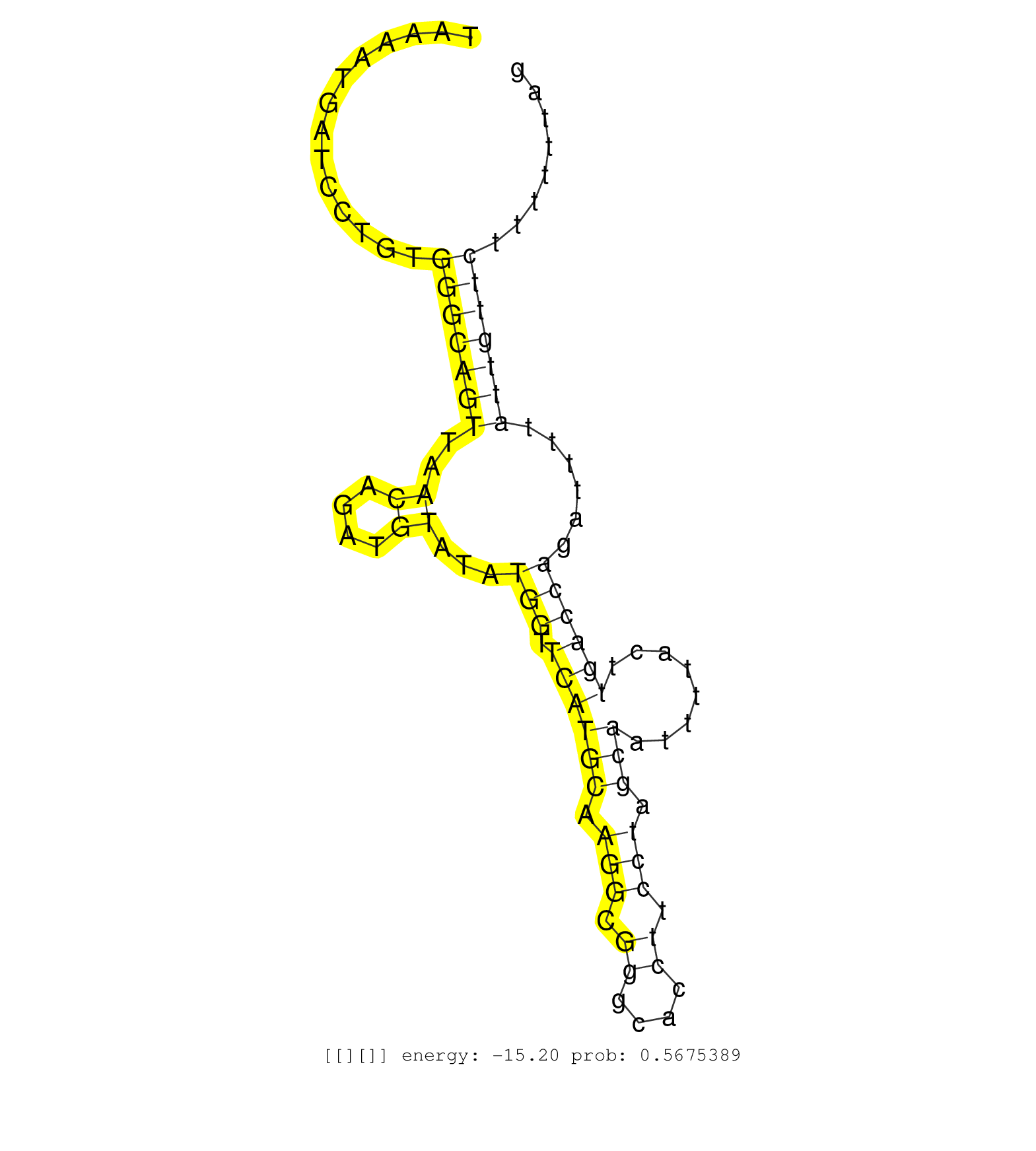
| TCTCATTAATTGTGGAAGCAACATATTGGATATCTGACTTCTTGTATCTTTTTGTTCTTGTATGGATGATTAAGATCTTCTTATACATGAAAATTGTATTTAAAATGATCCTGTGGGCAGTTAACAGATGTATATGGTTCATGCAAGGCGGGCACCTTCCTAGCAATTTTACTTGACCAGATTTTATTGTTCTTTTTTAGTGTGACCCAAATCTTCAGGTGTTTAGTGTTTTCATCGATAACCTTGATAC ..................................................................................................................(((((((..((....))...(((.((((((.(((.((....)).))).)))........))))))......))))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................TTCTTATACATGAAAATTGTATTTAAAATGATCCTGTGGGCAGTTAACAGAT......................................................................................................................... | 52 | 1 | 61.00 | 61.00 | 61.00 | - | - | - | - | - | - |
| .............................................................................................................................AGATGTATATGGTTtggt........................................................................................................... | 18 | tggt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 |
| TCTCATTAATTGTGGAAGCAACATATTGGATATCTGACTTCTTGTATCTTTTTGTTCTTGTATGGATGATTAAGATCTTCTTATACATGAAAATTGTATTTAAAATGATCCTGTGGGCAGTTAACAGATGTATATGGTTCATGCAAGGCGGGCACCTTCCTAGCAATTTTACTTGACCAGATTTTATTGTTCTTTTTTAGTGTGACCCAAATCTTCAGGTGTTTAGTGTTTTCATCGATAACCTTGATAC ..................................................................................................................(((((((..((....))...(((.((((((.(((.((....)).))).)))........))))))......))))))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................CAATTTTACTTGACCAtcca....................................................................... | 20 | tcca | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................ACTTGACCAGATTTTATTGTTCTTTTTTA................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| .............................CTGACTTCTTGTATgagt........................................................................................................................................................................................................... | 18 | gagt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................GCACCTTCCTAGCAgagc..................................................................................... | 18 | gagc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................CAAATCTTCAGGTGTTTAGTGTTTTCA................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |