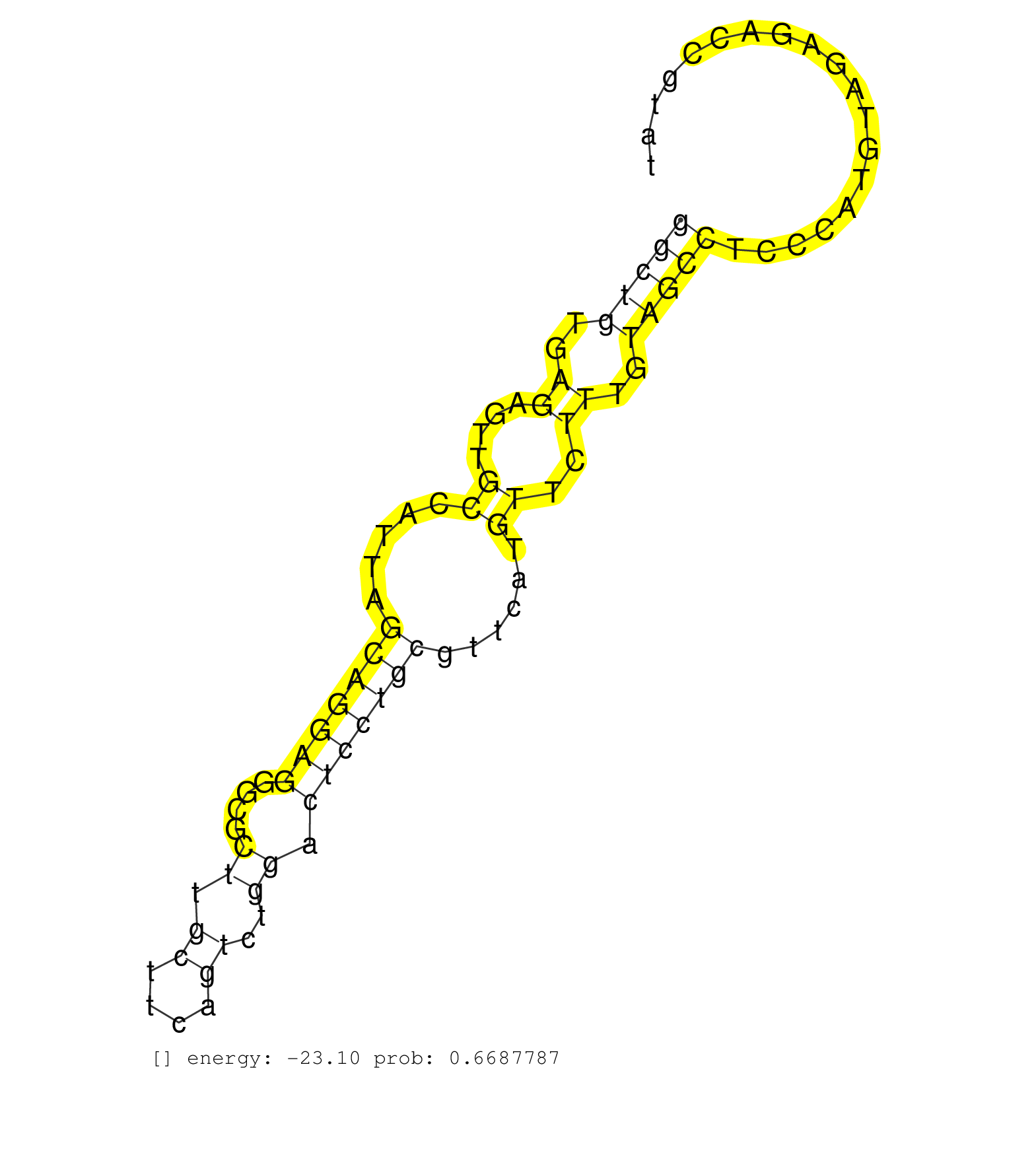

| Gene: Gpam | ID: uc008hxk.1_intron_1_0_chr19_55147261_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(6) PIWI.ip |
(17) TESTES |
| TGCTCCTCTCTCTGCTTTCTTTAATAGATGGCAGGATAGGTTGAAGAGATGGCAGGTCACTGGAAGAAGGCTGTGAGAGTTGCCATTAGCAGGAGGGCGCTTGCTTCAGTCTGGACTCCTGCGTTCATGTTCTTTGTAGCCTCCCATGTAGAGACCGTATGCTGAGCCTGAACACTGAAGGCCTTGTTGTCTCTCTGCAGCTGAGAGTGCCACATACTGTCTCGTGAAGAACGCTGTGAAAATGTTTAAG ....................................................................(((((..((....((.....(((((((....((.((....))..)).)))))))......))..))..)))))............................................................................................................. ....................................................................69.........................................................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................TGTTCTTTGTAGCCTCCCATGTAGAGACC.............................................................................................. | 29 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGCTGAGCCTGAACACTGAAGGCCTT................................................................. | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAGTGCCACATACTGTCTCGTGAA...................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................GTGCCACATACTGTCTCGTGA....................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAGTGCCACATACTGTCTCGTGA....................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................................AGAGACCGTATGCTGAGCCTGAACACTGAAG...................................................................... | 31 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCCCATGTAGAGACCGTATGCTGAGC................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGCAGCTGAGAGTGCCACATACTGT.............................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................GACCGTATGCTGAGCCTGAACA............................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................CCTCCCATGTAGAGACCGTATGCTGAGC................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GCTGAGCCTGAACACTGAAGGCCTTGT............................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................................................................................GAGTGCCACATACTGTCTCGTGAAGAACtt................ | 30 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................................TTGTAGCCTCCCATGTAGAGACCGTAT.......................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................ATACTGTCTCGTGAAGAACGCTGT............. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CCGTATGCTGAGCCTGAACACTGAAGGCC................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGTAGCCTCCCATGTAGAGACCGTAT.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGCTGAGCCTGAACACTGAAGGCCTTG................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TTCAGTCTGGACTCCTGCGTTCATGT........................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGTTCTTTGTAGCCTCCCATGTAGAGAC............................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGTAGAGACCGTATGCTGAGCCTGAAC............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCCTGCGTTCATGTTCTTTG.................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CGTATGCTGAGCCTGAACACTGAAGGC.................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAGAGACCGTATGCTGAGCCTGAAC............................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................ACCGTATGCTGAGCCTGAACACTGAAGGC.................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TGCTGAGCCTGAACACTGAAGGCCTTGT............................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................CTCCCATGTAGAGACCGTATGCTGAGCCT................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TTCTTTGTAGCCTCCCATGTAGAGACC.............................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCCCATGTAGAGACCGTATGCTGAGCC.................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TGAGAGTTGCCATTAGCAGGAGGGCGt...................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| TGCTCCTCTCTCTGCTTTCTTTAATAGATGGCAGGATAGGTTGAAGAGATGGCAGGTCACTGGAAGAAGGCTGTGAGAGTTGCCATTAGCAGGAGGGCGCTTGCTTCAGTCTGGACTCCTGCGTTCATGTTCTTTGTAGCCTCCCATGTAGAGACCGTATGCTGAGCCTGAACACTGAAGGCCTTGTTGTCTCTCTGCAGCTGAGAGTGCCACATACTGTCTCGTGAAGAACGCTGTGAAAATGTTTAAG ....................................................................(((((..((....((.....(((((((....((.((....))..)).)))))))......))..))..)))))............................................................................................................. ....................................................................69.........................................................................................160........................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|