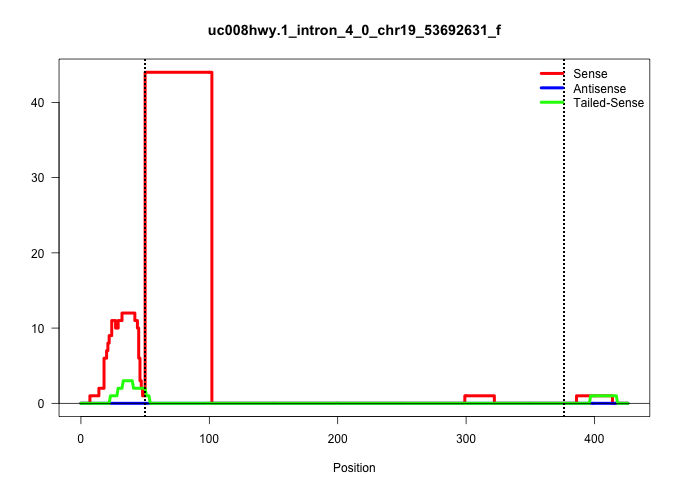
| Gene: Smc3 | ID: uc008hwy.1_intron_4_0_chr19_53692631_f | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(10) TESTES |
| TTTCTGCTTTTGTGGAAATCATTTTTGACAATTCGGACAACCGGTTACCAGTAAGTGACAGTCAATTTCTTTTCAGTAATGTTGAGAATTTTATGCAATAGCCTAAAATTATAAGCATTAAAGATTTATAATTTAGTTCATGCTAAAGTAAATAAGTAGTTTGTCAATGGATTTTGTATTTGAATTTTAATGAGCACTCTAGTGAACAGGATTATGTAAAGCAATTGTAAGAAAATGTTTTCTAAATATTATAGTATCATCTTAAGTGAAACCATTGGAAATCTGAAATAGTTATGATCAACGACTTTAAAATGAACTTAGGTAAAAGTTGTCCACCTGACCATCTGGGCTTTTTTCTTTTTTTTTTCCTTCTTAGATTGATAAAGAGGAAGTTTCACTTCGAAGAGTTATTGGTGCCAAAAAGGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTGACAGTCAATTTCTTTTCAGTAATGTTGAGAATTTTATGCAATAGC.................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 44.00 | 44.00 | 44.00 | - | - | - | - | - | - | - | - | - |
| ..................TCATTTTTGACAATTCGGACAACCGGT............................................................................................................................................................................................................................................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - |
| .......TTTTGTGGAAATCATTTTTG............................................................................................................................................................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................TTTGACAATTCGGACgcc................................................................................................................................................................................................................................................................................................................................................................................................. | 18 | gcc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................CTTCGAAGAGTTATTGGgtat........ | 21 | gtat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............GAAATCATTTTTGACAATTCGGACAACCGG.............................................................................................................................................................................................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................AATTCGGACAACCGGTTACCAattg.................................................................................................................................................................................................................................................................................................................................................................................... | 25 | attg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................TTTTGACAATTCGGACAACCGGTTA........................................................................................................................................................................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................AATTCGGACAACCGGTT............................................................................................................................................................................................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................TCATTTTTGACAATTCGGACAACCGGTT............................................................................................................................................................................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................AACGACTTTAAAATGAACTTAGG........................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................TTGACAATTCGGACAACC................................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................TTGACAATTCGGACAACCGGTT............................................................................................................................................................................................................................................................................................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................CGGACAACCGGTTACCAa....................................................................................................................................................................................................................................................................................................................................................................................... | 18 | a | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................ATTTTTGACAATTCGGACAACCGGT............................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................TCGGACAACCGGTTACCA........................................................................................................................................................................................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................TTTTTGACAATTCGGACAACCGGTTAC.......................................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................AGGAAGTTTCACTTCGAAGAGTTATTGG............ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| TTTCTGCTTTTGTGGAAATCATTTTTGACAATTCGGACAACCGGTTACCAGTAAGTGACAGTCAATTTCTTTTCAGTAATGTTGAGAATTTTATGCAATAGCCTAAAATTATAAGCATTAAAGATTTATAATTTAGTTCATGCTAAAGTAAATAAGTAGTTTGTCAATGGATTTTGTATTTGAATTTTAATGAGCACTCTAGTGAACAGGATTATGTAAAGCAATTGTAAGAAAATGTTTTCTAAATATTATAGTATCATCTTAAGTGAAACCATTGGAAATCTGAAATAGTTATGATCAACGACTTTAAAATGAACTTAGGTAAAAGTTGTCCACCTGACCATCTGGGCTTTTTTCTTTTTTTTTTCCTTCTTAGATTGATAAAGAGGAAGTTTCACTTCGAAGAGTTATTGGTGCCAAAAAGGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................GAATTTTATGCAATActgc...................................................................................................................................................................................................................................................................................................................................... | 19 | ctgc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |