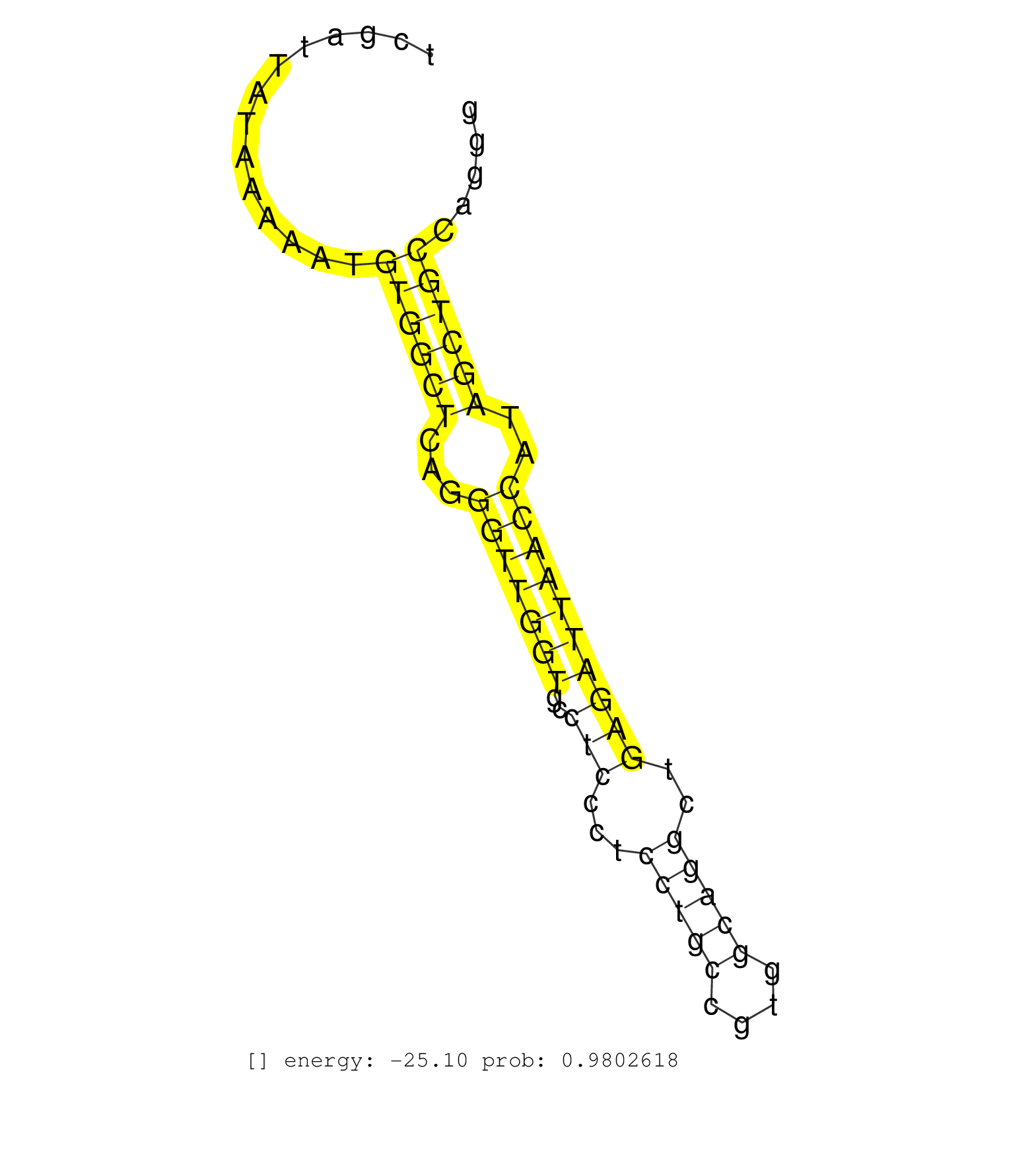
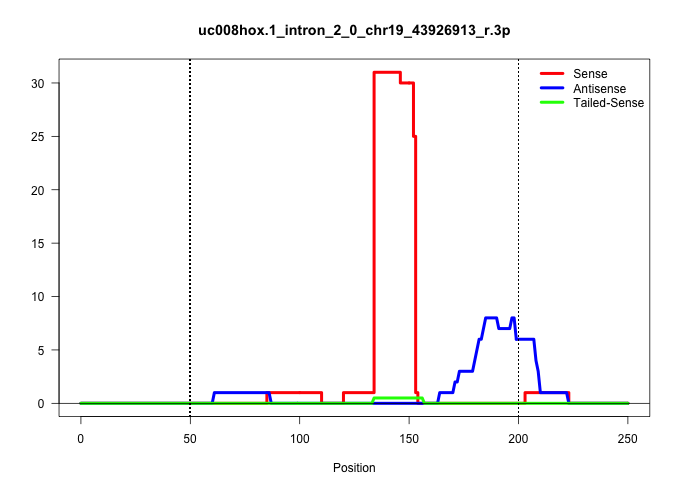
| Gene: Dnmbp | ID: uc008hox.1_intron_2_0_chr19_43926913_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(15) TESTES |
| TGTTGCATAAAGTAAGGGTGTTAGCTTGCCTCAGAGCCATTAAAACATTGCAGTGTGCTTTTTACCTAACACAGCCAGACTCGATTATAAAAATGTGGCTCAGGGTTGGTGCCTCCCTCCTGCCGTGGCAGGCTGAGATTAACCATAGCTGCCAGGGTGCCAGCGTGAGTTTTCTCATTGGCCCTGTTCTACCTTTTCAGCCAAGCTACATGCTGCAGTCGGAAGAGCTGCGGAGCTCCCTGCTGGCGAG ..............................................................................................((((((...(((((((..(((...(((((....)))))..))))))))))..)))))).................................................................................................. ................................................................................81..........................................................................157........................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................GAGATTAACCATAGCTGCC................................................................................................. | 19 | 2 | 24.00 | 24.00 | 16.50 | 7.00 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| ......................................................................................................................................GAGATTAACCATAGCTGC.................................................................................................. | 18 | 2 | 5.00 | 5.00 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................GAGATTAACCATAGCTGCCA................................................................................................ | 20 | 2 | 1.50 | 1.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...........................................................................................................................................................................................................AGCTACATGCTGCAGTCGGA........................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................TATAAAAATGTGGCTCAGGGTTGGT............................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................TGCCGTGGCAGGCTGAGATTAACCAT........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................GAGATTAACCATAGCTGCCAaaa............................................................................................. | 23 | aaa | 0.50 | 1.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| TGTTGCATAAAGTAAGGGTGTTAGCTTGCCTCAGAGCCATTAAAACATTGCAGTGTGCTTTTTACCTAACACAGCCAGACTCGATTATAAAAATGTGGCTCAGGGTTGGTGCCTCCCTCCTGCCGTGGCAGGCTGAGATTAACCATAGCTGCCAGGGTGCCAGCGTGAGTTTTCTCATTGGCCCTGTTCTACCTTTTCAGCCAAGCTACATGCTGCAGTCGGAAGAGCTGCGGAGCTCCCTGCTGGCGAG ..............................................................................................((((((...(((((((..(((...(((((....)))))..))))))))))..)))))).................................................................................................. ................................................................................81..........................................................................157........................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................CCTGTTCTACCTTTTCAGCCAAGCTA.......................................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................CCTAACACAGCCAGACTCGATTATAA................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................GTTCTACCTTTTCAGCCAAGCTACA........................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................TGTTCTACCTTTTCAGCCAAGCTACA........................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCTGTTCTACCTTTTCAGCCAAGCTACA........................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TTACCTAACACAGCCAGACTCGATTA................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTCTCATTGGCCCTGTTCTACCTTTTCA................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................TGGCAGGCTGAGATtagc............................................................................................................... | 18 | tagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................TACCTAACACAGCCAGACTCGATTA................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................GCCCTGTTCTACCTTTTCAGCCAAGCTA.......................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CCCTGTTCTACCTTTTCAGCCAAGCTAC......................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCCTGTTCTACCTTTTCAGCCAAGCTAt.......................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................GTGAGTTTTCTCATTGGCCCTGTTCTA........................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GCGTGAGTTTTCTCATTGGCCCTGTTCTA........................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CTCATTGGCCCTGTTCTACCTTTTCA................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGCCAAGCTACATGCTGCAGTCGGA........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |