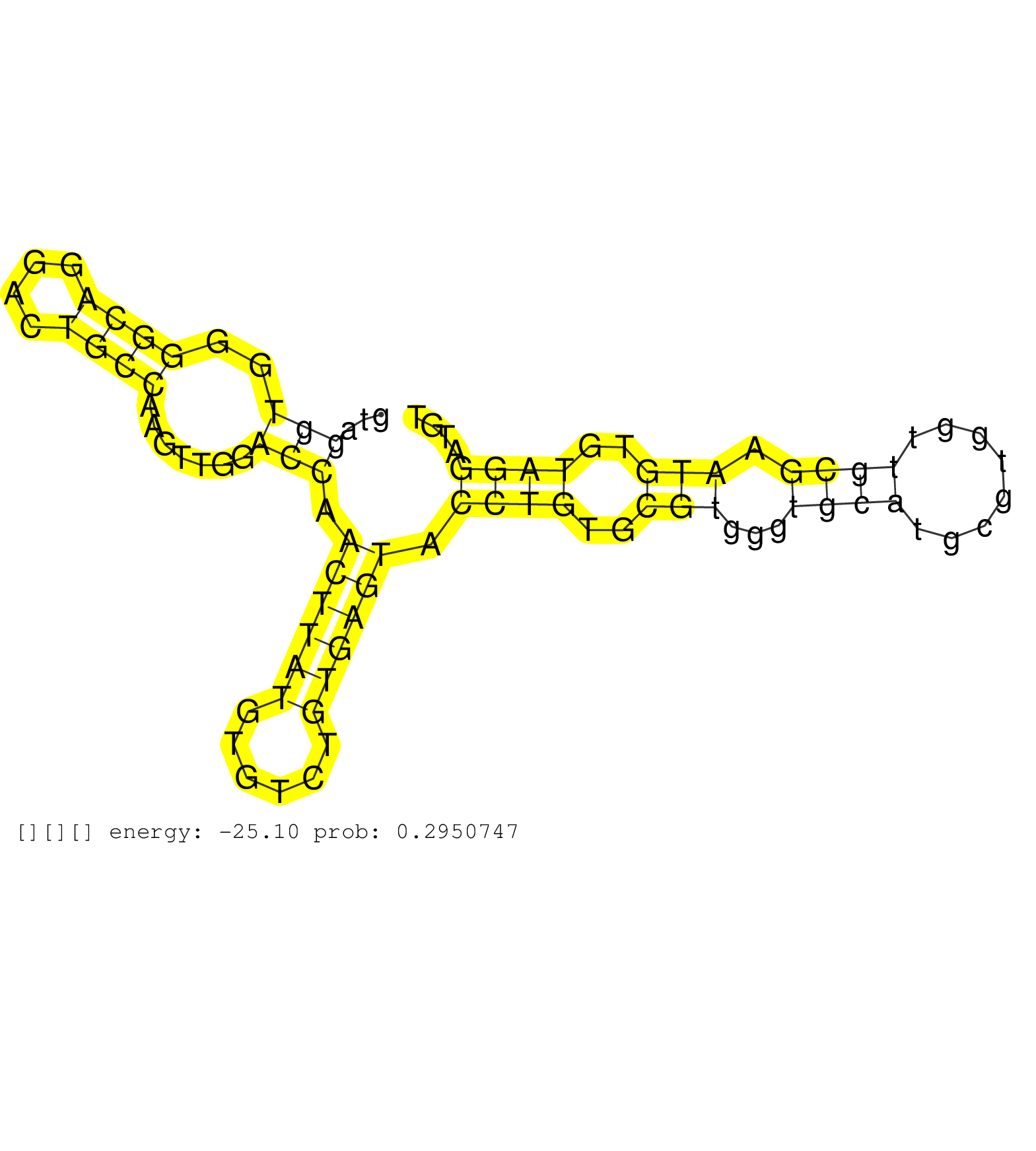

| Gene: Loxl4 | ID: uc008hns.1_intron_10_0_chr19_42679775_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(8) TESTES |
| GGGATGCTGGGCTTTCCCAGTCAGACATCTGTCAACAGCCACTACTACAGGTAGGTGGGGCAGGACTGCCAAGTTGGACCAACTTATGTGTCTGTGAGTACCTGTGCGTGGGTGCATGCGTGGTTGCGAATGTGTAGGATGTAAACCTTAAAGAGTTGCTCGTGTTTCTGTGTGTGTGTGTCGTGCGCAAGCACATGCACGCACTCTATAAATGCGCATGCGCAGTGTTGAATGGCTCGGTGAGATTCTG .....................................................(((..((((....)))).......))).((((((......)))))).((((..(((...((((........)))).)))..))))................................................................................................................ ..................................................51.........................................................................................142.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................TGGGGCAGGACTGCCAAGTTGGACCAACTTATGTGTCTGTGAGTACCTGTGC............................................................................................................................................... | 52 | 1 | 20.00 | 20.00 | 20.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................CGAATGTGTAGGATagg........................................................................................................... | 17 | agg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................................TGAATGGCTCGGTGAGgc.... | 18 | gc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - |
| GGGATGCTGGGCTTTCCCAGTCAGACATCTGTCAACAGCCACTACTACAGGTAGGTGGGGCAGGACTGCCAAGTTGGACCAACTTATGTGTCTGTGAGTACCTGTGCGTGGGTGCATGCGTGGTTGCGAATGTGTAGGATGTAAACCTTAAAGAGTTGCTCGTGTTTCTGTGTGTGTGTGTCGTGCGCAAGCACATGCACGCACTCTATAAATGCGCATGCGCAGTGTTGAATGGCTCGGTGAGATTCTG .....................................................(((..((((....)))).......))).((((((......)))))).((((..(((...((((........)))).)))..))))................................................................................................................ ..................................................51.........................................................................................142.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................TCTGTCAACAGCCACTACTACAGGTA..................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - |
| .....................CAGACATCTGTCAACAGCCACTACTA........................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - |
| ..........GCTTTCCCAGTCAGACATCTGTCAACA..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ..................AGTCAGACATCTGTCAACAGCCACTACTA........................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................TGTCGTGCGCAAGCAacaa......................................................... | 19 | acaa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 |
| ........................ACATCTGTCAACAGCCACTACTA........................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |