
| Gene: Dock8 | ID: uc008hbc.1_intron_17_0_chr19_25201654_f | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(7) TESTES |
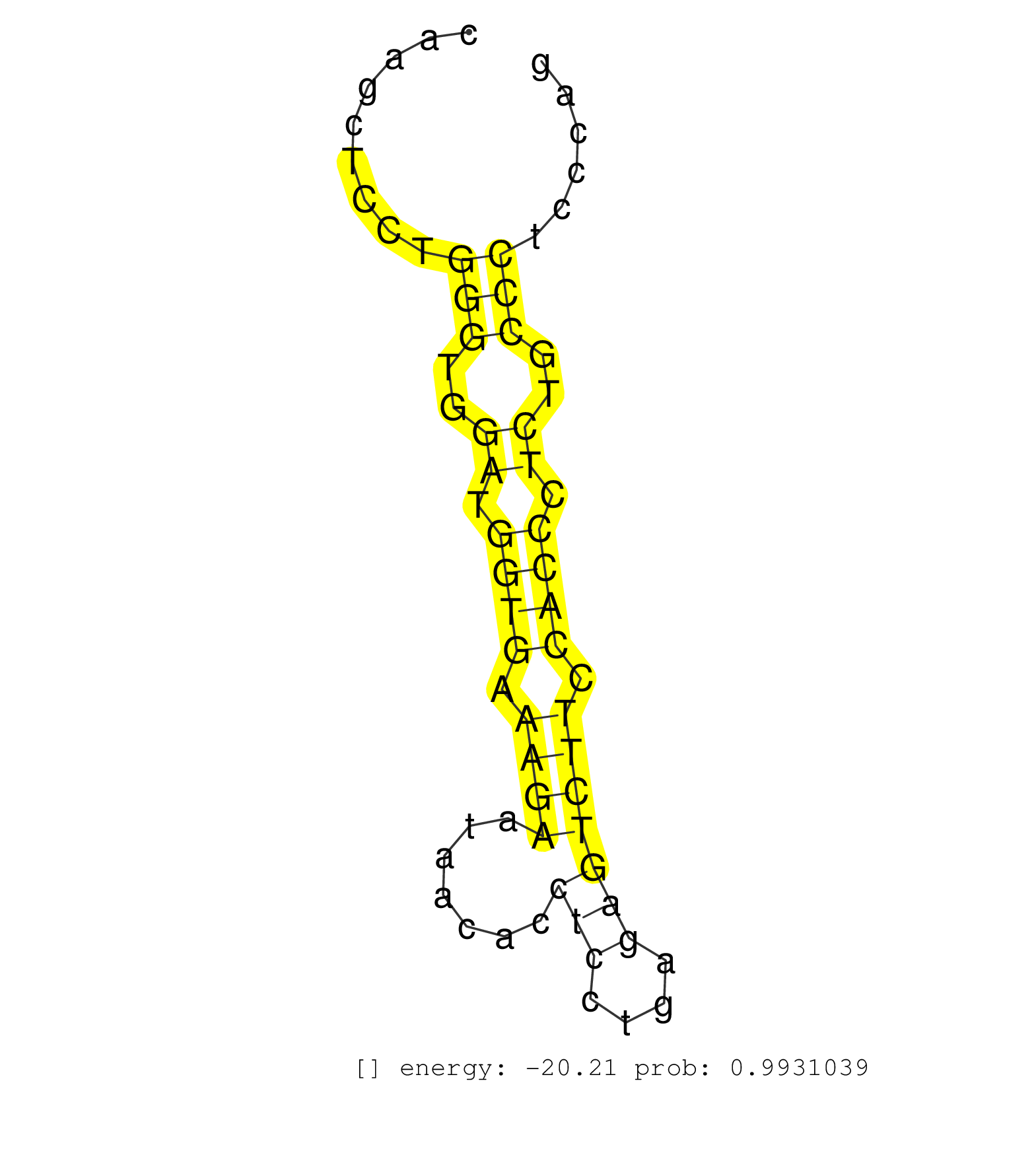
| AGGGGGTATTTAATATTGAAGTGCAAGCTGTTTCTTCCGTCCACACCCAGGTAAGGGACATCAAGGTCTGCCACCGTCCGGGTTAGCCATCTGCTTTGTGCTCTCTCTTGACAGTGTGATACCTAGTACATCACCTACCTACAGCATGAGTTGGAGCCTGGTTTATTGTTTCTTTAATTAATAGTTGTCAAAAGGTCCTATGTTGTATACCACATTCTACAAATAAGCCCACTGCCTTTGCTGGAAGCATCTGAGATTTCGAGTTCTAATCCAGAGCACTGGCTAAGGGATTGCACCAAGCTCCTGGGTGGATGGTGAAAGAATAACACCTCCTGAGAGTCTTCCACCCTCTGCCCTCCCAGGATAACCACCTGGAGAAGTTCTTCACCCTTTGCCACTCCCTGGAGAGCCA .................................................................................................................................................................................................................................................................................................................(((..((.((((.((((.......(((....))))))).)))).))..)))........................................................ ........................................................................................................................................................................................................................................................................................................297..............................................................362................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................................................................TCCTGGGTGGATGGTGAAAGA.......................................................................................... | 21 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................CCTGGGTGGATGGTGAAAGAAT........................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................GTCTTCCACCCTCTGCCC........................................................ | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................CCTGGGTGGATGGTGAAAGA.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - |
| ...........................................................................................TGCTTTGTGCTCTCaagt............................................................................................................................................................................................................................................................................................................... | 18 | aagt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................TACCTAGTACATCAtcct................................................................................................................................................................................................................................................................................... | 18 | tcct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 |
| AGGGGGTATTTAATATTGAAGTGCAAGCTGTTTCTTCCGTCCACACCCAGGTAAGGGACATCAAGGTCTGCCACCGTCCGGGTTAGCCATCTGCTTTGTGCTCTCTCTTGACAGTGTGATACCTAGTACATCACCTACCTACAGCATGAGTTGGAGCCTGGTTTATTGTTTCTTTAATTAATAGTTGTCAAAAGGTCCTATGTTGTATACCACATTCTACAAATAAGCCCACTGCCTTTGCTGGAAGCATCTGAGATTTCGAGTTCTAATCCAGAGCACTGGCTAAGGGATTGCACCAAGCTCCTGGGTGGATGGTGAAAGAATAACACCTCCTGAGAGTCTTCCACCCTCTGCCCTCCCAGGATAACCACCTGGAGAAGTTCTTCACCCTTTGCCACTCCCTGGAGAGCCA .................................................................................................................................................................................................................................................................................................................(((..((.((((.((((.......(((....))))))).)))).))..)))........................................................ ........................................................................................................................................................................................................................................................................................................297..............................................................362................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|