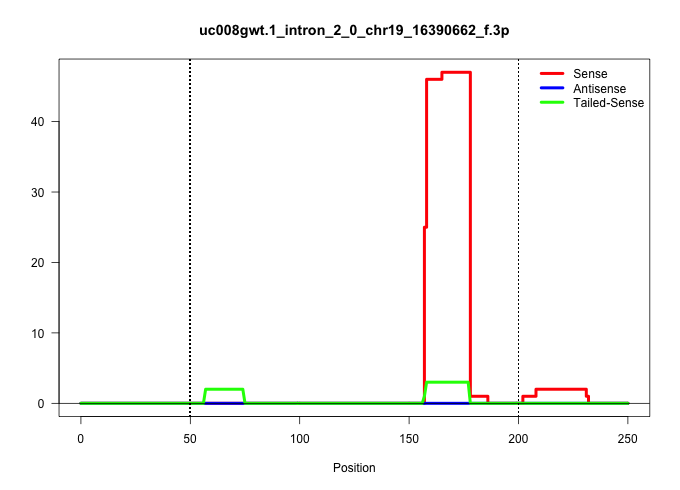
| Gene: Gnaq | ID: uc008gwt.1_intron_2_0_chr19_16390662_f.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(14) TESTES |
| GAGTTCAGCTGCTGAGGAAATGGGAATGTGGTAAGTAGGTGACTGGGTGGAGAACCTTTCTCTGAGCAGGGGAAGTGTGGCCTTGCCTGCTTCCTAGCCTGTCACCTTACAGACTTTCCTTTTTCTCTGTGACTCAGTTTTTGCTCTTTCCATGGGATTAGAATGATGGTAACTGACATTCTCATGTCTTCCTCTTCTAGCTATCTGAATGACTTGGACCGTGTAGCCGACCCTTCCTATCTGCCTACAC ......................................................(((......(((((.((((((((.(((...)))))))))))...(((.....(((((((............)))))))..)))....))))).......)))...((((((.(((...))).)))))).................................................................... ....................................................53...............................................................................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................TTAGAATGATGGTAACTGACA........................................................................ | 21 | 1 | 25.00 | 25.00 | 17.00 | 4.00 | - | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................TAGAATGATGGTAACTGACA........................................................................ | 20 | 1 | 21.00 | 21.00 | 15.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 |
| .........................................................TTCTCTGAGCAGGGctgt............................................................................................................................................................................... | 18 | ctgt | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TTAGAATGATGGTAACTGAta........................................................................ | 21 | ta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ATCTGAATGACTTGGACCGTGTAGCCGAC................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CTGAATGACTTGGACCGTGTAGCCG..................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................ATGGTAACTGACATTCTCATG................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................TATCTGAATGACTTGGACCGTGTA......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................ATGACTTGGACCGTGTAGCCGACC.................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TAGAATGATGGTAACTGAta........................................................................ | 20 | ta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TAGAATGATGGTAACTGACc........................................................................ | 20 | c | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GAGTTCAGCTGCTGAGGAAATGGGAATGTGGTAAGTAGGTGACTGGGTGGAGAACCTTTCTCTGAGCAGGGGAAGTGTGGCCTTGCCTGCTTCCTAGCCTGTCACCTTACAGACTTTCCTTTTTCTCTGTGACTCAGTTTTTGCTCTTTCCATGGGATTAGAATGATGGTAACTGACATTCTCATGTCTTCCTCTTCTAGCTATCTGAATGACTTGGACCGTGTAGCCGACCCTTCCTATCTGCCTACAC ......................................................(((......(((((.((((((((.(((...)))))))))))...(((.....(((((((............)))))))..)))....))))).......)))...((((((.(((...))).)))))).................................................................... ....................................................53...............................................................................................................................182.................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|