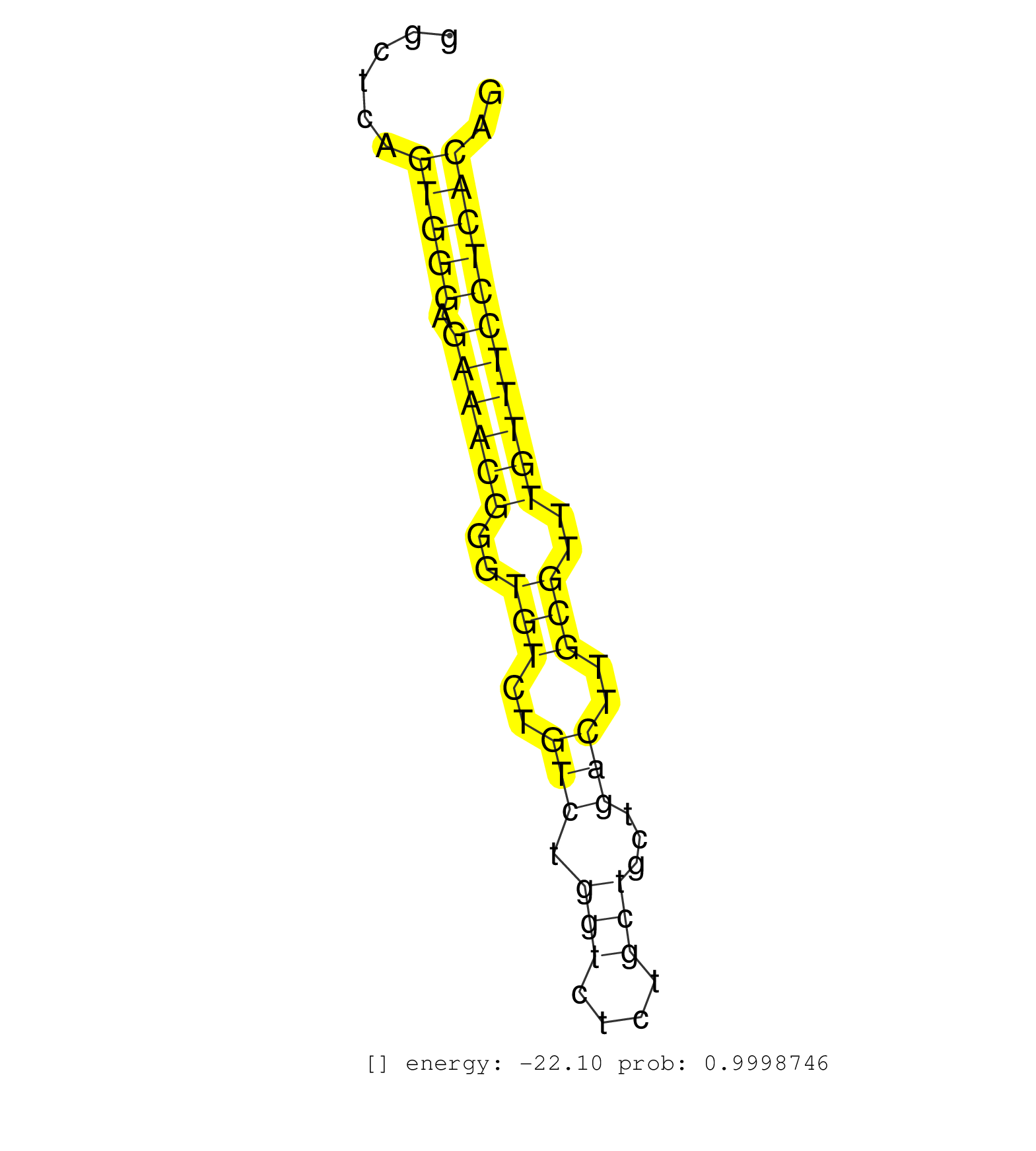

| Gene: Fads1 | ID: uc008gpe.1_intron_4_0_chr19_10261374_f.3p | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(1) PIWI.ip |
(4) PIWI.mut |
(22) TESTES |
| TCCTTGTCATGGCAGAGTCATAGGTCACAAGCCAGGGCCGGCCACTTGCTCCAGGAACATGTCAGAGACTTTCAAAGGTAGCAAAAAGCTAAAGGTTCTGGAAGTAGGCCTAGAGTCAGGACAGGGAAAGTTGGTGGCTCAGTGGGAGAAACGGGTGTCTGTCTGGTCTCTGCTGCTGACTTGCGTTTGTTTCCTCACAGCTCGGGAGGGAAAAGAAGAAGCACATGCCATACAACCATCAGCACAAGTA .............................................................................................................................................(((((.((((((..(((..(((.(((....)))...)))..)))..))))))))))).................................................... .......................................................................................................................................136.............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGT........................................................................................ | 22 | 1 | 7.00 | 7.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTC....................................................................................... | 23 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTt....................................................................................... | 23 | t | 3.00 | 7.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTTGCGTTTGTTTCCTCACAG.................................................. | 21 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................AGCTCGGGAGGGAAAAGAA................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTaa...................................................................................... | 24 | aa | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTat...................................................................................... | 24 | at | 1.00 | 7.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTCa...................................................................................... | 24 | a | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTCat..................................................................................... | 25 | at | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GGAAAGTTGGTGGagtg............................................................................................................. | 17 | agtg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTCT...................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTCaaag................................................................................... | 27 | aaag | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGcctg..................................................................................... | 25 | cctg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTttt..................................................................................... | 25 | ttt | 1.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTTGCGTTTGTTTCCTCACAGt................................................. | 22 | t | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................GACAGGGAAAGTTGGTGGCTCAGTGGGAGAAACGG................................................................................................ | 35 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CAGTGGGAGAAACGGGTGTCTGTC....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTtt...................................................................................... | 24 | tt | 1.00 | 7.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGTCTtaa................................................................................... | 27 | taa | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGGAGAAACGGGTGTCTGa........................................................................................ | 22 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................AGGACAGGGAAAGTTGGTGGCTCAGT........................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCTTGTCATGGCAGAGTCATAGGTCACAAGCCAGGGCCGGCCACTTGCTCCAGGAACATGTCAGAGACTTTCAAAGGTAGCAAAAAGCTAAAGGTTCTGGAAGTAGGCCTAGAGTCAGGACAGGGAAAGTTGGTGGCTCAGTGGGAGAAACGGGTGTCTGTCTGGTCTCTGCTGCTGACTTGCGTTTGTTTCCTCACAGCTCGGGAGGGAAAAGAAGAAGCACATGCCATACAACCATCAGCACAAGTA .............................................................................................................................................(((((.((((((..(((..(((.(((....)))...)))..)))..))))))))))).................................................... .......................................................................................................................................136.............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................GGCCGGCCACTTGCTCCAGGAACATGTCA.......................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GGTTCTGGAAGTAGGCCTAGAGTCAGGA................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |