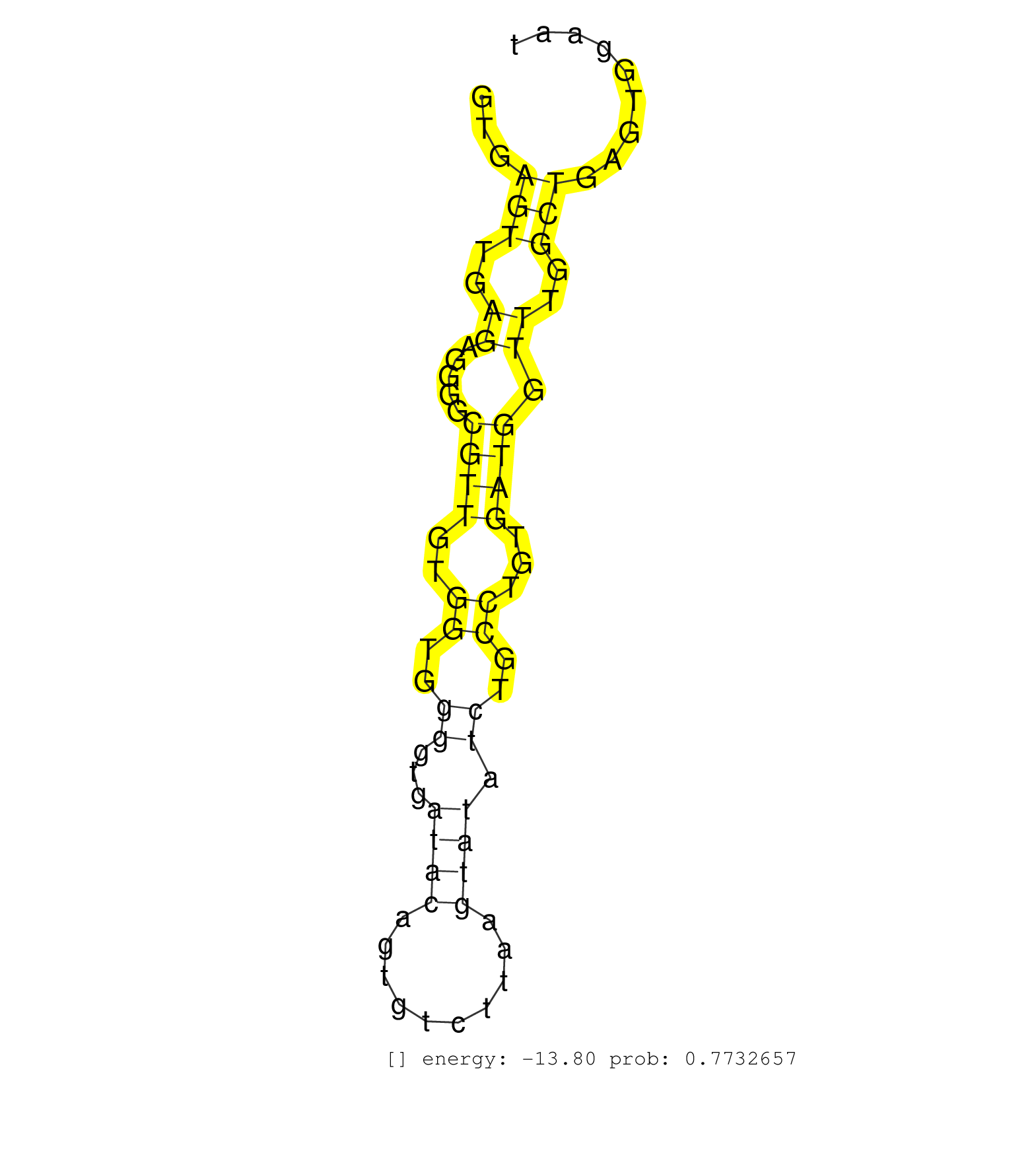

| Gene: Stx5 | ID: uc008gmo.1_intron_2_0_chr19_8817415_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(6) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(27) TESTES |
| CCGGCTCTGCACGCAGCCCGACAGTGCAGTGAATTTACCCTCATGGCCAGGTGAGTTGAGAGGGGCGTTGTGGTGGGGTGATACAGTGTCTTAAGTATATCTGCCTGTGATGGTTTGGCTGAGTGGAATTGGTTGTGTTGGAGGTTATGGTAACAGCCTCTTTCCTGCCCTTTACCACAACCTTCTCACTGATGATCATTTGCAGGCGCATTGGGAAAGATCTCAGCAATACATTTGCCAAGCTGGAGAAGCTAA .....................................................(((..((.....((((..((..((...((((..........)))).))..))...)))).))..)))....................................................................................................................................... ..................................................51............................................................................129............................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................TGCCTGTGATGGTTTGGCTGAGTG.................................................................................................................................. | 24 | 1 | 13.00 | 13.00 | 2.00 | - | 2.00 | 1.00 | - | 1.00 | 2.00 | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................TGCCTGTGATGGTTTGGCTGAGT................................................................................................................................... | 23 | 1 | 12.00 | 12.00 | 3.00 | - | - | 2.00 | - | 1.00 | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................TTCTCACTGATGATCATTTGCAG.................................................. | 23 | 1 | 9.00 | 9.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGAGAGGGGCGTTGTGGTG.................................................................................................................................................................................... | 25 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 |
| .....................................................................................................TGCCTGTGATGGTTTGGCTGAGTGGA................................................................................................................................ | 26 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGCCTGTGATGGTTTGGCTGAGTGGAA............................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TGCCTGTGATGGTTTGGCTGAGTGG................................................................................................................................. | 25 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGAGAGGGGCGTTGTGGT..................................................................................................................................................................................... | 24 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................GCCTGTGATGGTTTGGCTGAGT................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................CTGCCTGTGATGGTTTGGCTGAGTGGA................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TCTCACTGATGATCATTTGCAGa................................................. | 23 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................AAGCTGGAGAAGCTttgt | 18 | ttgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GCCTGTGATGGTTTGGCTGAGTGGA................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGATGGTTTGGCTGAGTGGAATTGGT.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TTCTCACTGATGATCATTTGCAGa................................................. | 24 | a | 1.00 | 9.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGAGAGGGGCGTTGTGG...................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TCTCACTGATGATCATTTGCAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGAGAGGGGCGTTGTGGTGc................................................................................................................................................................................... | 26 | c | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....CTCTGCACGCAGCCCGA.......................................................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GCCTGTGATGGTTTGGCTGAGTG.................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGAGAGGGGCGTTGTGGa..................................................................................................................................................................................... | 24 | a | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGTTGAGAGGGGCGTTGTGG...................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTTGAGAGGGGCGTTGTGGTaa................................................................................................................................................................................... | 26 | aa | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AGGCGCATTGGGAAAGATCTCA.............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCGGCTCTGCACGCAGCCCGACAGTGCAGTGAATTTACCCTCATGGCCAGGTGAGTTGAGAGGGGCGTTGTGGTGGGGTGATACAGTGTCTTAAGTATATCTGCCTGTGATGGTTTGGCTGAGTGGAATTGGTTGTGTTGGAGGTTATGGTAACAGCCTCTTTCCTGCCCTTTACCACAACCTTCTCACTGATGATCATTTGCAGGCGCATTGGGAAAGATCTCAGCAATACATTTGCCAAGCTGGAGAAGCTAA .....................................................(((..((.....((((..((..((...((((..........)))).))..))...)))).))..)))....................................................................................................................................... ..................................................51............................................................................129............................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................GGAAAGATCTCAGCAtgag........................... | 19 | tgag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |