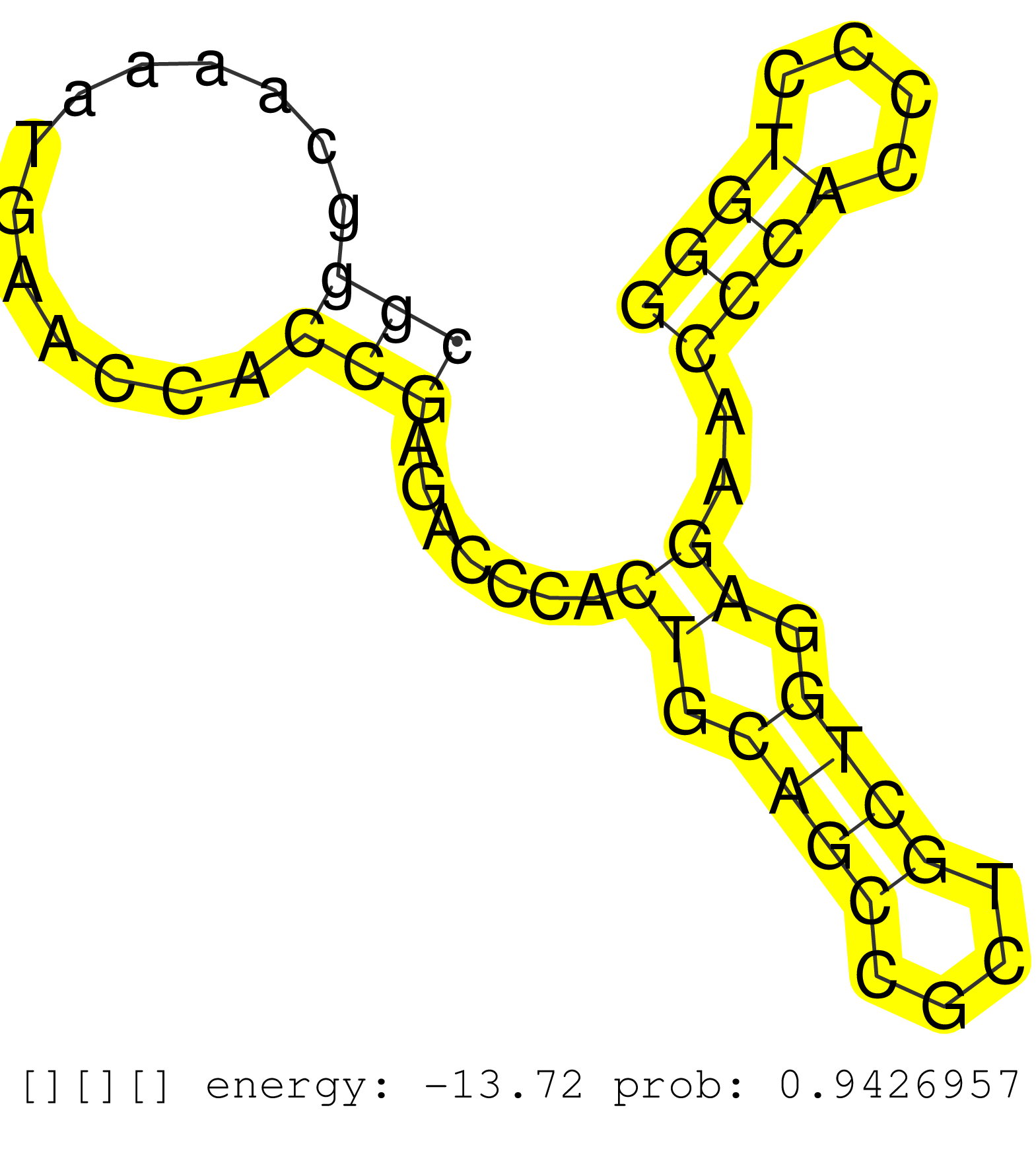

| Gene: Nrxn2 | ID: uc008giv.1_intron_21_0_chr19_6519328_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| CGCCCCCGGGGTGCTATTTGCGCCCTCCGCCCCAGCCCCCAACCTGCCCGCGGGCAAAATGAACCACCGAGACCCACTGCAGCCGCTGCTGGAGAACCCACCCCTGGGGCCTGGGGTCCCCACGGCCTTCGAGCCGCGGCGGCCGCCTCCCCTGCGCCCCGGCGTGACCTCAGCCCCCGGTTTCCCCCGTCTGCCCACAGCCAACCCCACGGGTCCGGGGGAGCGCGGCCCGCCAGGTGCAGTGGAGGTG ..................................................(((.............))).......((.((((....)))).))..((((....)))).............................................................................................................................................. ..................................................51.......................................................108............................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................TGAACCACCGAGACCCACTGCAGCCGC.................................................................................................................................................................... | 27 | 1 | 17.00 | 17.00 | 9.00 | 3.00 | - | 1.00 | 3.00 | 1.00 | - | - | - | - | - |
| ...........................................................TGAACCACCGAGACCCACTGCAGCCGCTGC................................................................................................................................................................. | 30 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - |
| ...........................................................TGAACCACCGAGACCCACTGCAGCCGCT................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ...........................................................TGAACCACCGAGACCCACTGCAGCCGCTG.................................................................................................................................................................. | 29 | 1 | 5.00 | 5.00 | - | - | 5.00 | - | - | - | - | - | - | - | - |
| ..........................................................ATGAACCACCGAGACCCACTGCAGCCG..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................CGGGCAAAATGAACCACCGAGACCCACTG........................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................TGCTGGAGAACCCACCCCTGGGGCCTG......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................AAATGAACCACCGAGACCCACTGCAGCC...................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................TGAACCACCGAGACCCACTGCAGCCG..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................AATGAACCACCGAGACCCACTGCAGCCGC.................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................TGAACCACCGAGACCCACTGCAG........................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CAACCCCACGGGTCCGtg............................... | 18 | tg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................TGAACCACCGAGACCCACTGCAGCCGCaa.................................................................................................................................................................. | 29 | aa | 1.00 | 17.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CGCCCCCGGGGTGCTATTTGCGCCCTCCGCCCCAGCCCCCAACCTGCCCGCGGGCAAAATGAACCACCGAGACCCACTGCAGCCGCTGCTGGAGAACCCACCCCTGGGGCCTGGGGTCCCCACGGCCTTCGAGCCGCGGCGGCCGCCTCCCCTGCGCCCCGGCGTGACCTCAGCCCCCGGTTTCCCCCGTCTGCCCACAGCCAACCCCACGGGTCCGGGGGAGCGCGGCCCGCCAGGTGCAGTGGAGGTG ..................................................(((.............))).......((.((((....)))).))..((((....)))).............................................................................................................................................. ..................................................51.......................................................108............................................................................................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................GCCGCCTCCCCTGCGCtgt............................................................................................. | 19 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................CCGGTTTCCCCCGTCTGCCCACAGCCAt............................................... | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CCTGGGGTCCCCACG.............................................................................................................................. | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | 0.25 | - |
| ..................................................................................................................................................................................................CCACAGCCAACCCCAC........................................ | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | 0.11 |