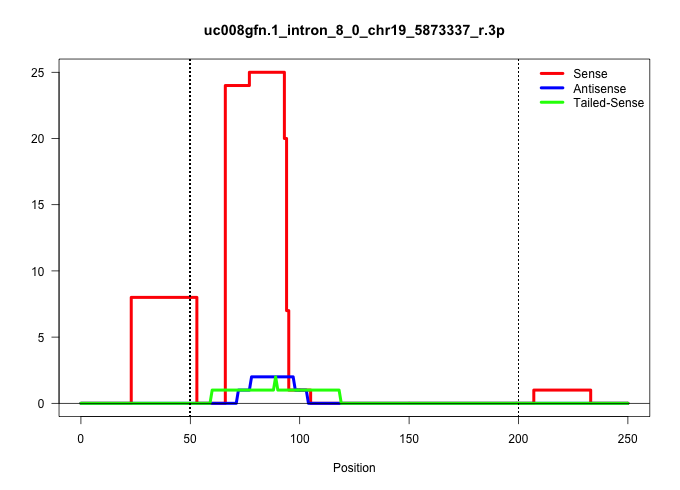
| Gene: 1200004M23Rik | ID: uc008gfn.1_intron_8_0_chr19_5873337_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(10) TESTES |
| TTGCTATGTAGCCCAGGCTGACCTGGGACTTGGAATCCTCCAGCTTTAGCCTCCTGCGTCCTGTCTTGGACTGCAGATGTATGTCACCGTAAGCCGTAGCGTCACGATGTGTCAGTGTGGCCTGGCTCGTGATAAGGAGCAAAGGGGTGGTGGGAAATGCTGATTGGACCTCACCCGACCCTGCCCGACCCGTATTCCAGCAGCCGATGTCCTGGTGTACCTGGCGGATGATACAGTAGTGCCCCTGGCC ....................................................................................................(((((((...(((((......))))))))))))..((.((..((((..((((((.....((....))..))))))...))))))))................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................TGGACTGCAGATGTATGTCACCGTAAGC............................................................................................................................................................ | 28 | 1 | 15.00 | 15.00 | 1.00 | 2.00 | 4.00 | 2.00 | 3.00 | 1.00 | 2.00 | - | - | - |
| .......................TGGGACTTGGAATCCTCCAGCTTTAGCCTC..................................................................................................................................................................................................... | 30 | 1 | 8.00 | 8.00 | 4.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................TGGACTGCAGATGTATGTCACCGTAAGCC........................................................................................................................................................... | 29 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................TGGACTGCAGATGTATGTCACCGTAAG............................................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - |
| .........................................................................................TAAGCCGTAGCGTCACGATGTGTCAGTGTt................................................................................................................................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................TGTATGTCACCGTAAGCCGTAGCGTCAC................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................TGTATGTCACCGTAAGCCGTAGCGTC................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................TGTCCTGGTGTACCTGGCGGATGATA................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................GTCTTGGACTGCAGATGTATGTCACCG................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................CTGTCTTGGACTGCAGATGTATGTCACaat................................................................................................................................................................ | 30 | aat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| TTGCTATGTAGCCCAGGCTGACCTGGGACTTGGAATCCTCCAGCTTTAGCCTCCTGCGTCCTGTCTTGGACTGCAGATGTATGTCACCGTAAGCCGTAGCGTCACGATGTGTCAGTGTGGCCTGGCTCGTGATAAGGAGCAAAGGGGTGGTGGGAAATGCTGATTGGACCTCACCCGACCCTGCCCGACCCGTATTCCAGCAGCCGATGTCCTGGTGTACCTGGCGGATGATACAGTAGTGCCCCTGGCC ....................................................................................................(((((((...(((((......))))))))))))..((.((..((((..((((((.....((....))..))))))...))))))))................................................................ ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................GCAGATGTATGTCACCGTAAGCCGTA........................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................GTATGTCACCGTAAGCCGTAGCGTCA.................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |