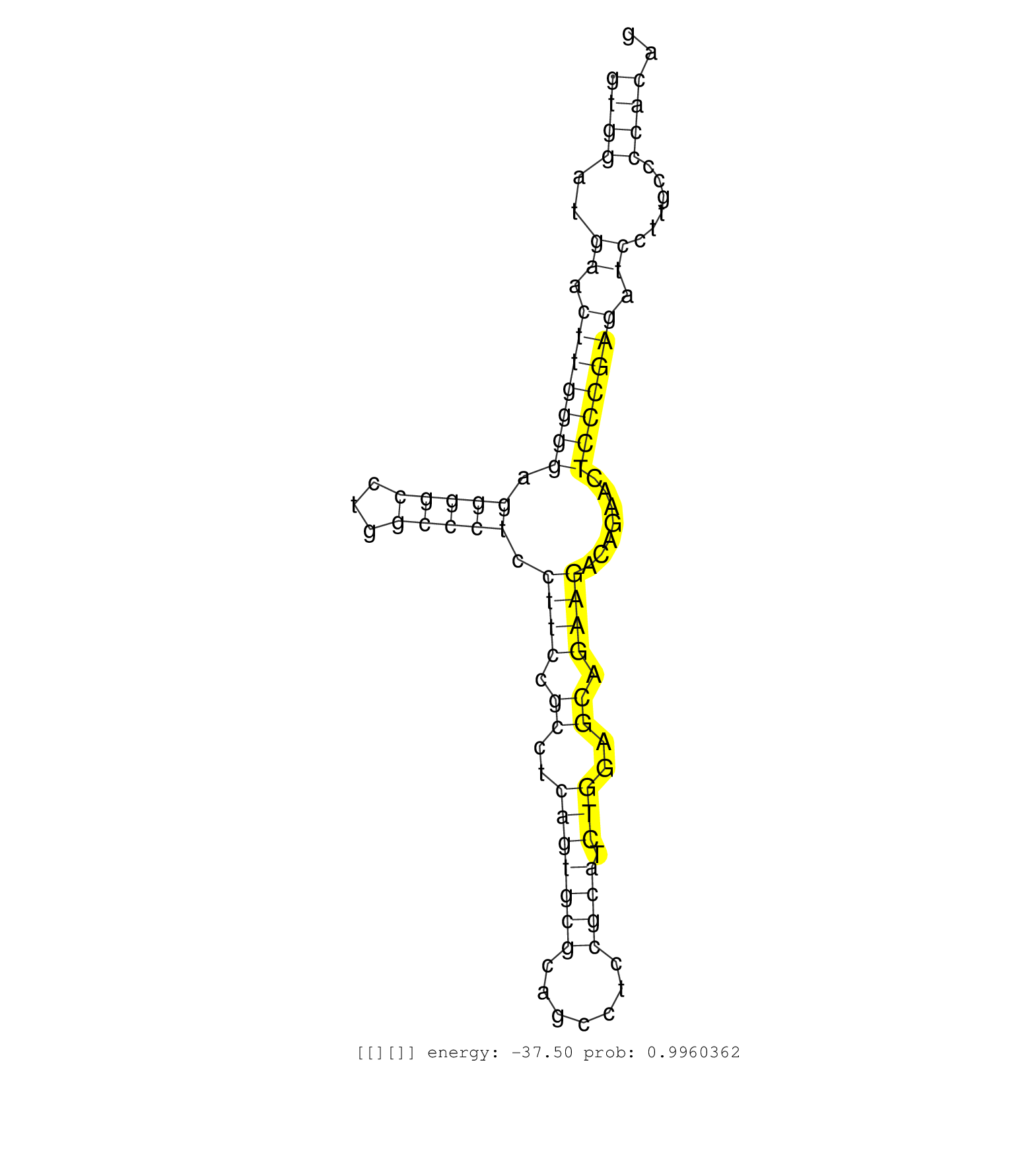

| Gene: Fibp | ID: uc008gdg.1_intron_6_0_chr19_5463851_f | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(26) TESTES |
| CCCTCCGGGGAAAGCTGGGTGTCTTCTCTGAGATGGAAACCAACTTCAAGGTCCGTTGCCCTGTGCAGACCCTTCCCCCTAAAGTGTCAAGAGCCTCCTGTGCGCTACCCCTGCCAGGCAGCTCTGCTCATTCCTGAAGGTGACCAGCAGGTGGCACTGCTGCTCTCCCTGTCTGCCCTTGAGGGGGTCCCAAGCTCATAACAAAAGGGACTTGATTGTCTGAGGGAGGTGGATGAACTTGGGGAGGGGCCTGGCCCTCCTTCCGCCTCAGTGCGCAGCCTCCGCATCTGGAGCAGAAGACAGAACTCCCGAGATCCTTGCCCCACAGAATCTGTCTCGGGGGCTGGTGAACGTGGCTGCCAAGCTGACCCACAATAA ....................................................................................................................................................................................................................................((((..((.(((((((.(((((...))))).((((.((..(((((((.......)))).)))..)).)))).......))))))).))......)))).................................................... ....................................................................................................................................................................................................................................229................................................................................................328................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................................................................................TCTGGAGCAGAAGACAGAACTCCCGA.................................................................. | 26 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................................................................................................................................................TCTGGAGCAGAAGACAGAACTCCCGAGA................................................................ | 28 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................TCTGGAGCAGAAGACAGAACTCCCGAG................................................................. | 27 | 1 | 4.00 | 4.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................TTCTCTGAGATGGAAACCAACTTCAAG........................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................CACAGAATCTGTCTCGGGGGCTGGTG............................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................GGGGCTGGTGAACGTGGCT.................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TAAAGTGTCAAGAGCCTCCTGTGCGC................................................................................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................TGGAGCAGAAGACAGAACTCCCGAGA................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................TCTGGAGCAGAAGACAGAACTCCCaaga................................................................ | 28 | aaga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GACTTGATTGTCTGAGGGAGGTGGAT................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TCTGAGGGAGGTGGATGAACTTGGG....................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................ATAACAAAAGGGACTTGATTGTCTGA........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................TGGAGCAGAAGACAGAACTCCCGAGATC.............................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................GACAGAACTCCCGAGATCCT............................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................GAATCTGTCTCGGGGGCTGGTGAACGTGG...................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................CTGTCTCGGGGGCTGGTGAACGTGGCTGC.................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AAAGCTGGGTGTCTTCTCTGA........................................................................................................................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................AACAAAAGGGACTTGATTGTCTGAG.......................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................TCTGGAGCAGAAGACAGAACTCCCGAGt................................................................ | 28 | t | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TCCGGGGAAAGCTGGGTGTCTTCTCT............................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................TGCCAGGCAGCTCTGCTCATTCCTGAA................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................AAGGGACTTGATTGTCTGAGGGAGGTG................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................GTCTCGGGGGCTGGTGAACGTGGCTGC.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................GAGATGGAAACCAACTTCAAGaat..................................................................................................................................................................................................................................................................................................................................... | 24 | aat | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TCTGCTCATTCCTGAAGGTG............................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................CGGGGGCTGGTGAACGTGGCTGCCAA............... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................GGATGAACTTGGGGAGG................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TCCCCCTAAAGTGTCAAGAGCCTCCT....................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................TCCTTGCCCCACAGAATCTGTCTCG....................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................TGAACGTGGCTGCCAAGCTGACCCACA.... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................TCTGTCTCGGGGGCTGGTGAACGTG....................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TGAGATGGAAACCAACTTCAAGGTCCGT.................................................................................................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................ACGTGGCTGCCAAGCTGA.......... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCTCCGGGGAAAGCTGGGTGTCTTCTCTGAGATGGAAACCAACTTCAAGGTCCGTTGCCCTGTGCAGACCCTTCCCCCTAAAGTGTCAAGAGCCTCCTGTGCGCTACCCCTGCCAGGCAGCTCTGCTCATTCCTGAAGGTGACCAGCAGGTGGCACTGCTGCTCTCCCTGTCTGCCCTTGAGGGGGTCCCAAGCTCATAACAAAAGGGACTTGATTGTCTGAGGGAGGTGGATGAACTTGGGGAGGGGCCTGGCCCTCCTTCCGCCTCAGTGCGCAGCCTCCGCATCTGGAGCAGAAGACAGAACTCCCGAGATCCTTGCCCCACAGAATCTGTCTCGGGGGCTGGTGAACGTGGCTGCCAAGCTGACCCACAATAA ....................................................................................................................................................................................................................................((((..((.(((((((.(((((...))))).((((.((..(((((((.......)))).)))..)).)))).......))))))).))......)))).................................................... ....................................................................................................................................................................................................................................229................................................................................................328................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................GTGCGCTACCCCtgg........................................................................................................................................................................................................................................................................... | 15 | tgg | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................GCTCTGCTCATTCCTGAAGGTGACCA........................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................................................GCCCTTGAGGGGGTCCCAAGCTCATAACA............................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGCCAGGCAGCTCTGCTCATTCCTGA................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................CTGAGATGGAAACCAAtagg............................................................................................................................................................................................................................................................................................................................................... | 20 | tagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................CAGGCAGCTCTGCTCATTCCTGAAGGTGA........................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |