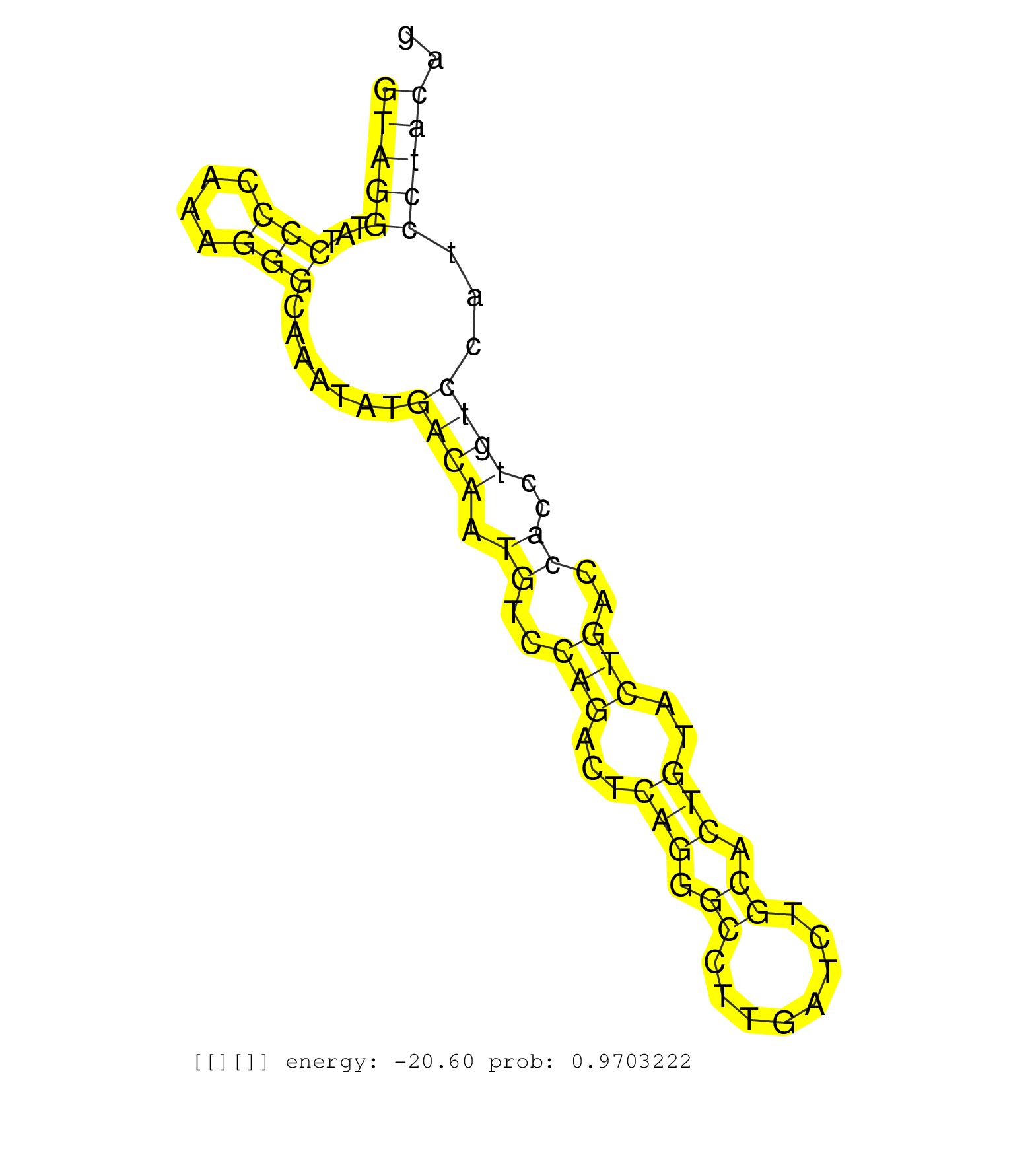

| Gene: Coro1b | ID: uc008fyy.1_intron_4_0_chr19_4150622_f | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(7) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| AGGACAAGAGCGTCCGAATCATCGATCCTAGGCGGGGCACCCTGGTGGCAGTAGGTATCCCCAAAGGGCAAATATGACAATGTCCAGACTCAGGGCCTTGATCTGCACTGTACTGACCACCTGTCCATCCTACAGGAACGTGAAAAGGCTCACGAGGGGGCCCGGCCCATGCGGGCCATCTTTCT ..................................................(((((...(((....))).......((((.((..(((...(((.((........)).)))..)))..))..))))...))))).................................................... ..................................................51..................................................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAGGTATCCCCAAAGGGCAAATATGACAATGTCCAGACTCAGGGCCTTGAT................................................................................... | 52 | 1 | 78.00 | 78.00 | 78.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TCCGAATCATCGATCCTAGGCGGGGCA.................................................................................................................................................. | 27 | 1 | 5.00 | 5.00 | - | 1.00 | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TACAGGAACGTGAAAAGGCTCACGAGG............................ | 27 | 1 | 4.00 | 4.00 | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................CATCGATCCTAGGCGGGGCA.................................................................................................................................................. | 20 | 1 | 3.00 | 3.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TCCGAATCATCGATCCTAGGCGGGGCAC................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GAGCGTCCGAATCATCGATCCTAGGC........................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................TATGACAATGTCCAGACTCAGGGCCT....................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGGTGGCAGTAGGTATCCCCAAAGGGC.................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TCCGAATCATCGATCCTAGGCGGGGCACC................................................................................................................................................ | 29 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCCTACAGGAACGTGAAAAGGCTCAC................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TGGTGGCAGTAGGTATCCCCAAAGGG..................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TACAGGAACGTGAAAAGGCTCACGAG............................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................TGTCCAGACTCAGGGCCTTGATCTGCACT............................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CCCCAAAGGGCAAATtgt............................................................................................................. | 18 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...ACAAGAGCGTCCGAATCATCGATCCTAGGC........................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................CCTAGGCGGGGCACCCTGGTGGCAGaac................................................................................................................................... | 28 | aac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................ATCCTAGGCGGGGCAta................................................................................................................................................ | 17 | ta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................GTACTGACCACCTGTCCAT......................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................TGTCCATCCTACAGGAACGTGAAAAGGCT................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TGCACTGTACTGACCACCTGTC............................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...ACAAGAGCGTCCGAATCATCGATCCTA........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................TCCTAGGCGGGGCACCCTGGTGGCAGaacg.................................................................................................................................. | 30 | aacg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......GAGCGTCCGAATCATCGATCCTAGGCG....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................ATCTGCACTGTACTGAC.................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| AGGACAAGAGCGTCCGAATCATCGATCCTAGGCGGGGCACCCTGGTGGCAGTAGGTATCCCCAAAGGGCAAATATGACAATGTCCAGACTCAGGGCCTTGATCTGCACTGTACTGACCACCTGTCCATCCTACAGGAACGTGAAAAGGCTCACGAGGGGGCCCGGCCCATGCGGGCCATCTTTCT ..................................................(((((...(((....))).......((((.((..(((...(((.((........)).)))..)))..))..))))...))))).................................................... ..................................................51..................................................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAAGAGCGTCCGgggc......................................................................................................................................................................... | 16 | gggc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................GGGCAAATATGACAATGTCCAGACTCA............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |